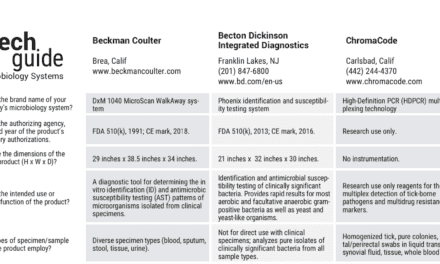
Metagenomic sequencing (mNGS) is a powerful diagnostic tool to detect causative pathogens in clinical microbiological testing. Rapid and accurate classification of metagenomic sequences is a critical procedure for pathogen identification in the dry-lab step of mNGS tests. However, this crucial step may be improved by classifying sequences within a clinically relevant timeframe – using GPMeta.
To address this challenge, a BGI Genomics team led by Xuebin Wang has recently launched GPMeta, an ultra-fast pathogen detection approach, and published these highlights in Briefings in Bioinformatics.
GPMeta can quickly and accurately identify pathogens through complex and massive mNGS sequencing data. Using simulated datasets and metagenomic sequencing datasets from clinical samples, results were benchmarked against tools used by the bioinformatics research community such as Bowtie2, Bwa, Kraken2, and Centrifuge.
Results show that GPMeta not only has higher accuracy but also exhibits significantly faster speed. In addition, GPMeta offers a GPMetaC clustering algorithm, a statistical model for clustering and re-scoring ambiguous alignments to improve the discrimination of highly homologous sequences from microbial genomes with average nucleotide identity >95%. These results underline GPMeta’s key role in the development of the mNGS test in infectious diseases that require rapid turnaround times.
GPMeta is a powerful tool to identify pathogens from mNGS data in a timely and accurate manner, which is of great importance in eliminating the threat from severe acute infections and in targeting precise and effective antibiotic therapy.
Moreover, GPMeta supports multiple GPUs to perform alignment and taxonomic classification of microbial sequences on split databases simultaneously and automatically merges results from multiple sub-databases, which is significant to keep up with the rapidly expanding microbial genome database. To make the best use of GPMeta, how to best and easily integrate it into clinical practices needs further study.