New whole exome and transcriptome sequencing assay demonstrates high concordance with existing companion diagnostics while requiring minimal tissue input for biomarker detection.
A new study validated the analytical and clinical performance of a US Food and Drug Administration (FDA)-approved test called MI Cancer Seek, an assay used as a companion diagnostic (CDx) to identify cancer patients who may benefit from targeted therapies.
It includes one pan-cancer and five tumor-specific indications for numerous FDA-approved therapies. MI Cancer Seek combines whole exome sequencing and whole transcriptome sequencing with FDA-approved CDx indications for solid tumor profiling in both adult and pediatric patients.
The test, developed by Caris Life Sciences, demonstrated over 97% agreement with other FDA-approved companion diagnostics in clinical comparisons, according to research published in Oncotarget on August 13. The assay successfully detects genetic alterations including PIK3CA, EGFR, BRAF, and KRAS/NRAS mutations while measuring tumor mutational burden (TMB) and microsatellite instability (MSI).
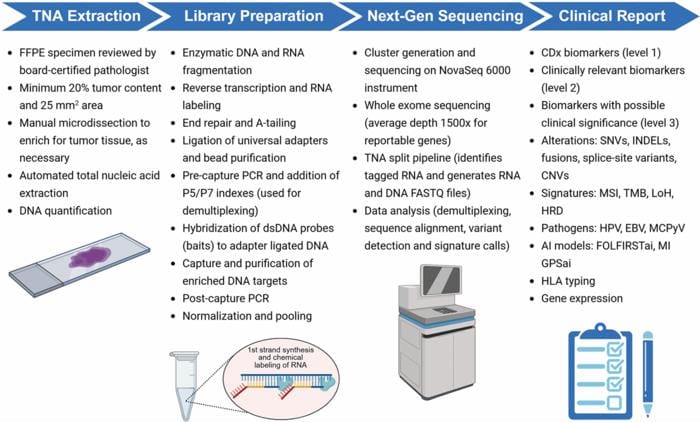
MI Cancer Seek requires only 50 ng of tissue input and maintains high accuracy levels even when analyzing formalin-fixed paraffin-embedded samples. The test workflow begins with total nucleic acid extraction from FFPE tissue slides requiring minimum 20% tumor content and 25 mm² area, obtained through manual microdissection if needed.
“MI Cancer Seek provides a comprehensive molecular blueprint that saves tissue without compromising results. The study results underscore our commitment to ongoing scientific integrity and validation,” says David Spetzler, MS, PhD, MBA, president of Caris, in a release. “Patients and physicians deserve this level of diligence and scientific rigor when selecting a molecular profiling assay to inform cancer treatment.”
Technical Performance and Clinical Applications
The assay showed near-perfect accuracy in identifying MSI status in colorectal and endometrial cancers and maintained precision across different laboratory conditions and varying DNA input levels. Sequencing is performed on qualified Illumina NovaSeq 6000 instruments, with raw data processed through Caris’ proprietary bioinformatics pipeline.
The test identifies biomarkers linked to FDA-approved treatments for breast, lung, colon, melanoma, and endometrial cancers. Beyond companion diagnostic capabilities, MI Cancer Seek incorporates features for detecting homologous recombination deficiency, structural variants, and cancer-related viruses.
Additional analytical tools include a Genomic Probability Score for identifying tissue of origin in cancers of unknown primary and a gene signature to guide first-line chemotherapy in colorectal cancer. The generated report includes companion diagnostic biomarkers with level 1 evidence, clinically relevant biomarkers with level 2 evidence, and biomarkers with possible clinical significance as level 3 evidence.
The validation study, led by first authors Valeriy Domenyuk and Kasey Benson, along with Spetzler, confirmed the test’s robustness for routine clinical laboratory use. However, researchers noted one limitation: low positive percent agreement for ERBB2 copy number alteration detection.
The assay’s ability to deliver multiple biomarker results from minimal tissue input addresses a key challenge for laboratories processing degraded FFPE samples. This efficiency could help laboratories streamline diagnostic workflows while reducing testing costs and connecting patients to targeted therapies more quickly.
As precision medicine continues expanding, the test provides laboratories with a single-platform solution for meeting increasing demands for molecular profiling in oncology diagnostics. The FDA approval enables clinical laboratories to implement the assay as part of their molecular testing menu for cancer biomarker detection.
Photo caption: MI Cancer Seek begins with total nucleic acid (TNA) extraction from formalin-fixed, paraffin-embedded (FFPE) tissue slides. A minimum 20% tumor content and 25 mm2 is required, which is obtained through manual microdissection if required. During library preparation, RNA is labeled during first strand cDNA synthesis. Sequencing is performed on qualified Illumina NovaSeq 6000 instruments. Raw data is processed by Caris’ proprietary bioinformatics pipeline, and a report is generated that includes CDx biomarkers (level 1 evidence), clinically relevant biomarkers (level 2 evidence), and biomarkers with possible clinical significance (level 3 evidence). Abbreviations: CNV: copy number variation; EBV: Epstein-Barr virus; GPS: genomic probability score; HLA: human leukocyte antigen; HPV: human papilloma virus; HRD: homologous recombination deficiency; INDEL: insertion/deletion; LoH: loss of heterozygosity; MCPyV: Merkel cell polyomavirus; MSI: microsatellite instability; SNV: single nucleotide variant; TMB: tumor mutational burden. Created in BioRender. Ribeiro, J. (2025) https://BioRender.com/m29z318.