Summary:
A study on Neuroblastoma with bone or bone marrow metastasis (NB-BBM) used deep learning and single-cell transcriptomics to uncover predictive biomarkers and potential therapeutic targets, advancing risk assessment and understanding of tumor progression.
Takeaways:
- A Swin-Transformer deep learning model predicted NB-BBM occurrence with over 85% accuracy using histopathological images.
- Single-cell transcriptomics identified tumor and macrophage subpopulations linked to NB-BBM progression, with oxidative phosphorylation playing a crucial role.
- Immune checkpoint genes (CD274, LAG3, TIGIT) were significantly upregulated, suggesting potential immunotherapy targets, while TKT was identified as a key metabolic molecule in NB-BBM malignancy.
Neuroblastoma (NB), the most prevalent extracranial solid tumor among children, is characterized by a high rate of metastasis. The pathogenesis of NB with bone or bone marrow metastasis (NB-BBM) and its complex immune microenvironment remain poorly understood, posing challenges for effective risk prediction for BBM and limiting therapeutic strategies.
This research, published in the Genes & Diseases journal by a team from The Children’s Hospital of Chongqing Medical University, highlights key genomic and single-cell transcriptomic alterations in NB-BBM, underscoring the significance of predictive pathology for NB-BBM and its role in understanding tumor onset, progression, and heterogeneity.
Neuroblastoma Study Uses Deep Learning to Predict Cancer Occurrence
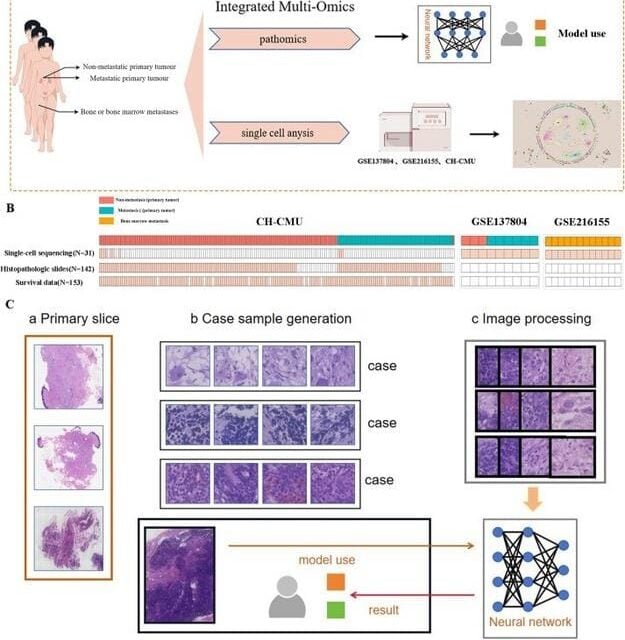
The researchers used a Swin-Transformer deep learning model to analyze 142 paraffin-embedded hematoxylin-eosin-stained tumor section images to predict neuroblastoma-BBM occurrence, achieving a classification accuracy exceeding 85%. In parallel, single-cell transcriptomics identified a tumor cell subpopulation (NB3) and two tumor-associated macrophage (TAM) subpopulations (SPP1+ TAMs and IGHM+ TAMs) closely associated with BBM progression. Interestingly, findings reveal that oxidative phosphorylation (OXPHOS) also plays a crucial role in BBM development.
Additionally, this study highlighted transketolase (TKT) as a crucial metabolic molecule linked to BBM. The researchers showed that the TKT gene was strongly associated with the clinical features of NB patients, especially in the BBM group. Functional experiments validated TKT’s involvement in malignant behavior, while pathway enrichment analysis showed correlations between high TKT expression and cell cycle activity.
Potential Targets Suggested
Moreover, expression analysis of immune checkpoint genes CD274, LAG3, and TIGIT revealed their significant upregulation in Neuroblastoma-BBM, suggesting potential targets for antibody-based immunotherapies. Furthermore, immunohistochemical validation demonstrated a pronounced expression of PD-L1 in NB-BBM, indicating its potential as a biomarker.
Although this research provides a predictive model for NB-BBM risk assessment, it has certain limitations, including the need for multicenter validation of the predictive model and prospective studies to confirm clinical utility. Despite these challenges, this study offers a pathodiagnostic prediction for the risk of NB-BBM, enhances other imaging diagnoses, and elucidates the cellular heterogeneity of initial, progressive, and distant metastatic sites in neuroblastoma.
Featured Image: (A) Experimental scheme for investigating the mechanisms underlying NB-BBM. (B) The sample overviews available for single-cell, whole genome sequencing, pathohistological, and survival data. (C) Workflow of a deep learning model for a multi-instance learning framework to predict NB-BBM. Image: Genes & Diseases