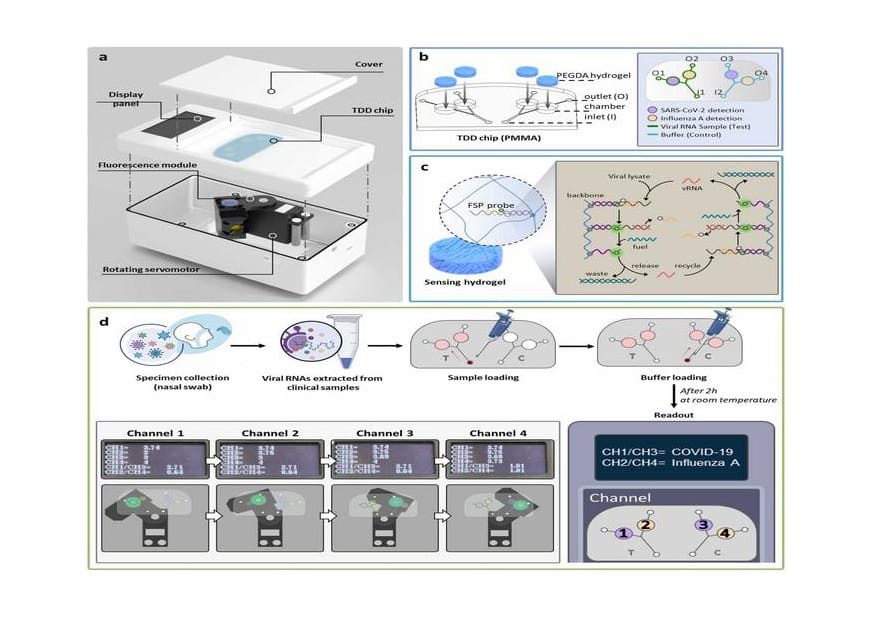
Summary: The TwinDemic Detection (TDD) system is a cost-effective, rapid, and sensitive diagnostic tool that detects SARS-CoV-2 and influenza A virus simultaneously at the point of care.
Key Takeaways:
- Advanced Diagnostic Technology: The TDD system uses a transparent microfluidic chip with hydrogel-based, enzyme-free gene detection sensors for precise viral detection.
- High Diagnostic Accuracy: The TDD demonstrated 93.3% accuracy for positive COVID-19 cases and 100% for influenza A cases in nasopharyngeal swab testing.
- Scalability and Potential: The system’s modular design allows for future enhancements, enabling detection of a broader range of viruses for pandemic preparedness.
The COVID-19 outbreak in 2019 triggered measures to raise public awareness regarding pandemics and also led to fast-tracked vaccine development. While these measures helped reduce viral transmission significantly, it also had some unintended consequences, such as an overall reduction in the spread of other viruses, leading to pauses in vaccination regimes. However, fast-mutating pathogens like viruses still pose a significant threat, with predictions of co-infections with multiple viruses causing “twindemics” or “tripledemics” in the future.
Reverse Transcriptase-quantitative PCRs (RT-qPCRs) are reliable disease diagnostic assays but are constrained by the use of expensive equipment and reagents, limiting their utility in resource-constrained settings. Therefore, a rapid, accurate, and sensitive molecular diagnostic tool is warranted for the simultaneous detection of multiple viruses at the point-of-care.
The TwinDemic Detection (TDD) System for COVID and Influenza Testing
To address this gap, a team of scientists from the Republic of Korea, led by Professor Eunjung Kim from Incheon National University (INU), recently developed a novel TwinDemic Detection (TDD) system, designed for simultaneous detection of SARS-CoV and influenza A virus (IAV). Their findings were published in the Sensors and Actuators B: Chemical journal.
“The TDD includes a transparent poly (methyl methacrylate) microfluidic chip with hydrogel-based, enzyme-free gene detection sensors, along with a handheld fluorescence reader,” says Kim as she describes the detection system. The hydrogel chambers are embedded with customized probes to detect the two target viral pathogens: SARS-CoV-2 (CoV) and IAV. The reaction between the target viral DNA and the specific probe system amplifies the fluorescence signal.
Notably, the TDD system is easy to use, cost-effective, and has a detection limit of 0.46 picomolar (pM) for CoV and 0.39 pM for IAV. To confirm TDD’s diagnostic efficiency, 15 nasopharyngeal swabs each from healthy individuals, patients with COVID-19, and those with Flu A were tested. For COVID-19 diagnosis, the TDD system correctly predicted positive samples in 93.3% of the cases, and negative samples in 96.7% of the cases. For IAV, positive and negative samples were correctly predicted in 100% and 96.7% of the cases, respectively.
“The application of our TDD system can be further expanded by introducing additional channels and sensing hydrogels on the microfluidic chip, as well as integrating highly sensitive nucleic acid amplification systems for simultaneous detection and differentiation of a wider range of viruses,” says Kim, elaborating on the prospects of the TDD system against the team’s findings.
Overall, this study presents and highlights the TDD system as a novel point-of-care diagnostic tool that enables an accurate and rapid on-site detection of multiple viruses simultaneously and could aid clinicians make timely and appropriate treatment decisions.
Featured image: Simultaneous infections by multiple respiratory viruses are challenging to diagnose. Against this backdrop, scientists from Incheon National University have now developed a rapid, accurate, and sensitive point-of-care assay for detecting SARS-CoV-2 and influenza A virus simultaneously, aiding in limiting viral transmissions. Photo: Prof. Eunjung Kim from INU, Korea. Image source link: https://doi.org/10.1016/j.snb.2024.136933