The next-generation sequencing (NGS) module added to the Omics Explorer program from bioinformatics software company Qlucore, Lund, Sweden, comprises an interactive genome browser and interactive filter options. The module synchronizes expression analysis of RNA-seq data and genomic information, helping clinical researchers to improve their productivity.
Qlucore Omics Explorer enables researchers and clinicians to interactively explore and analyze small to large datasets, including data generated with NGS technologies. The software combines powerful statistical methods with real-time visualization. Such a combination shortens analysis time, adds more creativity to the research process, strengthens the path to new findings, and facilitates easier and more fruitful collaboration among biologists and bioinformaticians.

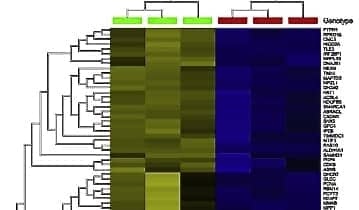
Omics Explorer hierarchical clustering and heatmaps of 1,747 differentially expressed RNA transcripts (A) and 336 proteins (B) in scramble control (SC Image 1) and ALDH1A1 knock down (KD Image 2) shRNA-transfected COLO320 cells.
Qlucore Omics Explorer provides a rapid understanding of any data problem, including those with up to many thousands of changing features. Users can rapidly visualize correlations in the data and understand how data samples might cluster and relate to one another. Such an overview can then provide clues for further investigation.
Founded in 2007 by researchers in the departments of mathematics and clinical genetics at Lund University, Qlucore has customers in about 25 countries around the world. Many of the leading pharmaceutical companies, hospitals, and universities around the world use Qlucore in their research.
For more information visit Qlucore.