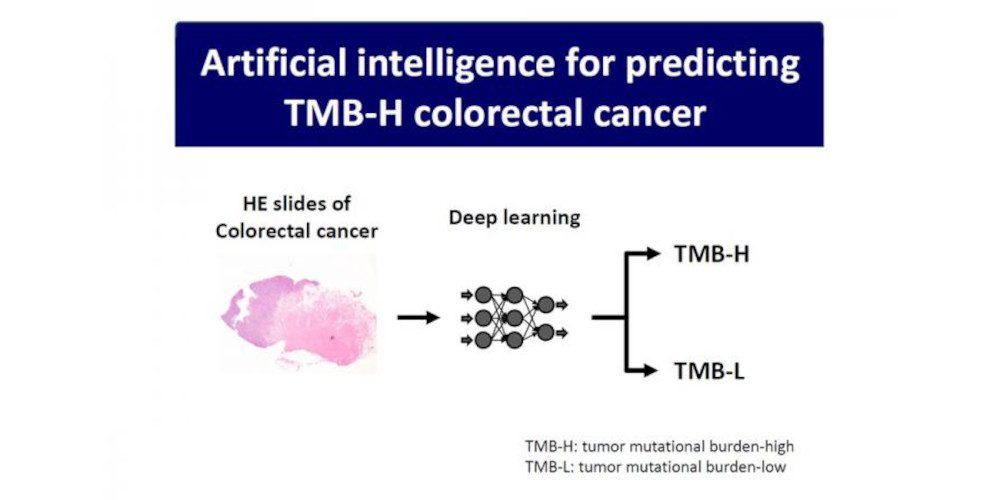
In a new study published in the Journal of Gastroenterology, researchers at Niigata University in Japan describe how they were able to use artificial intelligence to help identify genetic alterations in TMB-H colorectal cancer.
Biomarkers are important determinants of appropriate and effective therapeutic approaches for various diseases including cancer. There is ample evidence pointing to the significance of immune checkpoint inhibitors (ICI) against cancer, and they showed promising clinical benefits to a specific group of patients with colorectal cancer (CRC). Several reports demonstrated the efficacy of biomarkers such as programmed death-1 protein ligand (PD-L1), density of tumor-infiltrating lymphocytes (TILs), and tumor mutational burden (TMB), to determine the patient responsiveness for the efficient use of ICIs as therapeutics against cancer.
A high level of TMB (TMB-H), which reflects elevated total number of non-synonymous somatic mutations per coding area of a tumor genome and normally derived from gene panel testing, is recognized as a promising biomarker for the ICI therapies of various solid cancers. However, in clinical practice, it is not feasible to perform gene panel testing for all cancer patients.
Yoshifumi Shimada, MD, PhD, and coworkers from the Division of Digestive and General Surgery, Graduate School of Medical and Dental Sciences, Niigata University, determined TMB-H from a specific CRC patient subgroup to be a more robust marker for predicting the efficacy of ICIs, and developed a convolutional neural network (CNN)-based algorithm to predict TMB-H CRC directly from the histopathological characteristics (in particular, the TIL) obtained from the hematoxylin and eosin (H&E)-stained slides.
A representative microscopic image of the H&E-stained tumor mutational burden-high colorectal cancer tumor is shown in the accompanying figure, demonstrating the presence of tumor-infiltrating lymphocytes in significantly elevated levels compared to that of the normal surrounding tissue. Digital information from such neoplastic and also non-neoplastic images obtained from JP-CRC-cohort is transformed and normalized to build a predictive Convoluted Neural Network model by employing the Inception V3 learning model from Shimada’s group.
The CNN-based model developed by Shimada and his coworkers has the potential to not only reduce the burden on pathologists to properly diagnose patients, but also to provide the necessary information about patients’ responsiveness to ICI-based therapeutics, without the use of expensive, time consuming, and not easily available gene panel testing.
In addition, the studies completed by Shimada’s group provided means to predict TMB-H CRC using only the TIL information from the H&E slides from the patients’ tumor tissues. However, considering that the patients in the studied cohort were not treated with any ICIs, no conclusions could be drawn regarding their ICI responsiveness following the TMB-H diagnosis and it was suggested that future clinical trials be conducted to address whether TIL alone can be useful as a predictive biomarker for the efficacy of ICIs.
“We have developed artificial intelligence to predict genetic alterations in colorectal cancer by deep learning using hematoxylin and eosin slides,” Shimada says. “This artificial intelligence is important in solving the cost problems associated with genetic analysis and facilitating personalized medicine in colorectal cancer.”
Overall, the studies by Shimada and his associates provide a reliable and cost- and time-effective method to inform clinicians if the CRC patients they are managing can benefit from immune checkpoint inhibitor therapy (including inhibitors of the PD-1 protein and its ligand, PD-L1), without necessitating the use of a gene panel.
Featured image: A representative microscopic image of the hematoxylin and eosin (H&E) stained tumor mutational burden-high colorectal cancer tumor. Digital information from such neoplastic and also non-neoplastic images is transformed and normalized to build a predictive convoluted neural network (CNN) model employing the Inception V3 learning model. Credit: Niigata University