Where we are now, and what comes next
By Albino Troilo, PhD
The unprecedented public health crisis caused by covid-19 has led to illness, death, and worldwide economic disaster. Symptoms of severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) infection range from none at all to mild fevers and coughs, to severe pneumonia and multi-organ failure.1 From the beginning, diagnostic testing—to determine if a patient has an active or a past infection—has been critical to monitoring and treating patients and controlling the spread of this disease.
Currently, there are two general types of covid-19 tests:
Diagnostic tests are used to determine whether a patient has an active infection and needs to be quarantined to prevent further spread of the disease. Most of these are molecular tests, which are generally categorized as nucleic acid amplification tests (NAATs) because they detect viral RNA in patients’ samples, usually from nasopharyngeal swab specimens. Molecular tests rely on reverse transcription (RT) of the viral RNA into DNA, followed by polymerase chain reaction (PCR) amplification of the DNA and subsequent detection. Most frequently, the detection is based on reverse-transcription quantitative PCR (RT-qPCR).
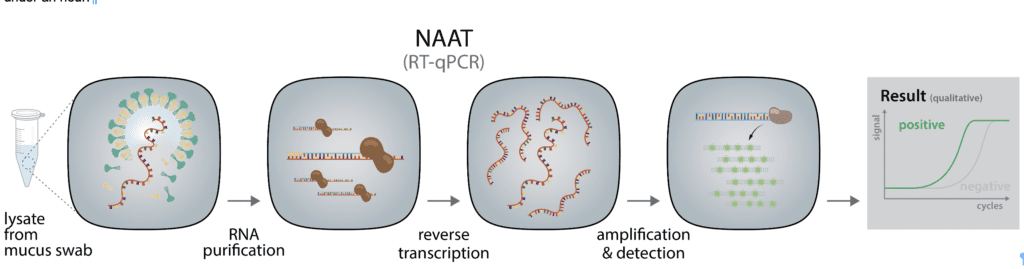
The underlying principle of these RT-qPCR-based tests is simple (Figure 1): Specific genetic sequences are used to detect the virus. A nasal or throat swab is performed to harvest viral particles and virus-infected mucosa cells. The swab sample is then lysed and the viral RNA is extracted and reverse-transcribed into cDNA. The cDNA in the processed sample is then quantified by qPCR to detect the presence of the virus’s genome and therefore confirm infection. Different test methods use varying steps to obtain the same result; automatization and optimization of sample preparation and reverse transcription can reduce the time it takes to get results from several days to under an hour. Molecular tests are reliable and accurate and are performed by specialized clinical laboratories.
Antigen tests, which detect specific proteins on the surface of the virus, are an alternative diagnostic for covid-19. These tests detect viral antigens in nasal or throat swab samples using antibody-based assays against specific SARS-CoV-2 viral proteins. Although these tests are specific for the virus and faster and less expensive than molecular tests, they aren’t as sensitive as PCR tests. For a positive result, antigen tests require the presence of a considerable amount of viral proteins in the specimen samples,2 which increases the chances of false negative results in low-grade infected patients.3 For this reason, negative results from an antigen test may need to be confirmed with a PCR test prior to making treatment decisions.
Antibody tests detect antibodies that are produced by the immune system in response to an infection. While molecular tests serve as the frontline diagnostic tools to monitor and combat the covid-19 pandemic, antibody tests, also called immunogenic serology tests, are used to detect antibodies produced by the patient’s immune system in response to a prior infection. Serological detection of SARS-CoV-2 antibodies determines whether patients have been exposed to SARS-CoV-2, even if they are currently asymptomatic.
Antibodies—immunoglobulins categorized as IgM, IgA, and IgG—can take several days or weeks to develop after an infection and may stay in the blood for several weeks or more after recovery. IgM is released as a pentameric antibody and is generally among the first responses of the humoral immune system. IgA is secreted as a dimeric antibody and plays a major role in the defense of the mucosal epithelia of the respiratory airways and the intestine. IgG typically appears later and becomes a major component of the immune memory response and immunity. Seroconversion, which is the time period needed until specific antibodies are detectable in the blood, is typically within the first 2 weeks.4-7
Due to the delay between infection, symptomatic onset, and the detectable presence of antibodies, serological tests should not be used to diagnose active infection. However, they can be used to confirm past infection in recovered patients or to screen asymptomatic patients. Additionally, these tests can aid in the better characterization of the disease, including: understanding the kinetics of the immune response to infection; understanding the immune response relative to disease severity and timeline; clarifying whether cross-reactivity with other coronaviruses leads to cross-protection; understanding whether past infection protects from future infection and how long immunity will last; determining the correlates of protection that can guide public health measures; selecting potential blood donors of convalescent plasma, which may serve as a possible treatment for seriously ill covid-19 patients; and seroepidemiological studies to understand the extent of covid-19’s spread.
Three different types of antibody tests can be used for covid-19:
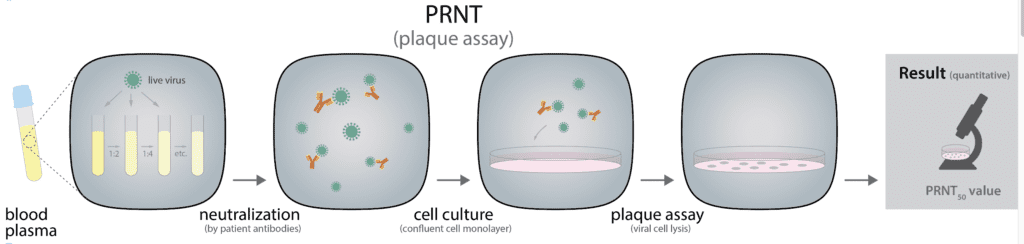
• Plaque reduction neutralization test (PRNT) is performed in a laboratory setting and identifies antibodies that can neutralize a viral infection (Figure 2). PRNT requires whole blood, serum, or plasma from the patient. The sample is diluted, mixed, and incubated with a viral suspension to allow the antibodies to react with the antigens present on the surface of the virus. Following the incubation period, the solution is poured on a layer of host cells that allow for the growth of SARS-CoV-2 virus. The host cells are then covered with a layer of agar or cellulose to prevent viral spreading, and localized plaques (infected regions) form after a few days. The plaques are then measured by microscopy observation or fluorescent dyes that react with the infected cells. Plaque formation is affected by the presence of neutralizing antibodies in the patient’s serum. The concentration of serum to reduce the number of plaques by 50% compared to a control sample without serum provides the PRNT50 value, which is the measure of how much antibodies are present in the sample and how effective they are.
PRNT is considered to be the gold standard for detecting and measuring neutralizing antibodies for a specific virus.8,9 However, due to the test’s complexity and slow turnaround time, it is not suitable for high-throughput screenings.
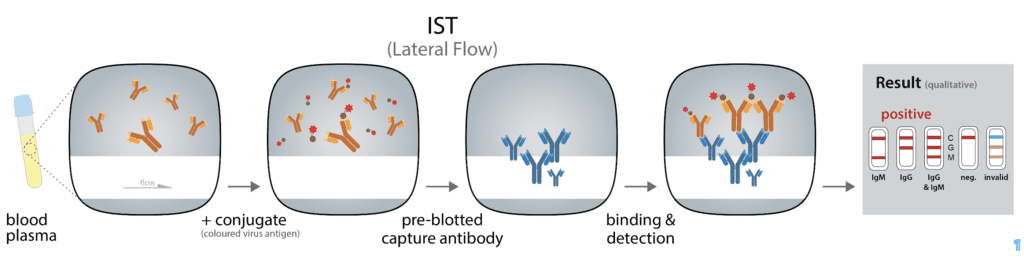
• Immunochromographic strip test (IST) is a qualitative (positive or negative) lateral flow assay that is small, portable, and affordable, and can be used in point-of-care settings. Often performed using a few blood drops from a finger prick, this rapid diagnostic test (RDT) looks like a common pregnancy test (Figure 3). ISTs rely on immobilized antibodies and detection with colloidal gold-conjugated
SARS-CoV-2 antigens. Colloidal gold is composed of very small gold particles (5-100nm), which are an intense red color. A few drops of blood, serum, or plasma are added onto a sample pad and passed over a detection strip by capillary tension. As it moves, the sample passes a conjugate/reagent pad, where it is mixed with the conjugated viral antigens. When the blood contains antibodies that bind with the viral antigen-conjugate, an antigen-antibody complex is formed. The sample-conjugate mix then passes over detection stripes, zones where anti-human IgG or IgM antibodies have been placed. Here, the antibodies contained in the sample will be immobilized, and if they are bound to conjugated viral antigens, the detection stripe will stain red. By having independent stripes of anti-IgM and anti-IgG antibodies, both subclasses of virus-specific antibodies can be detected individually. A positive control consisting of an antibody of a different species and the respective conjugate is included to indicate that the test was carried out correctly.
Point-of-care RDTs offer ease of use and rapid results, but they are intrinsically hampered by the short incubation times defined by the capillary flow, the comparably small sample amounts, and the lack of wash steps, which limits the sensitivity and specificity of the assay type.
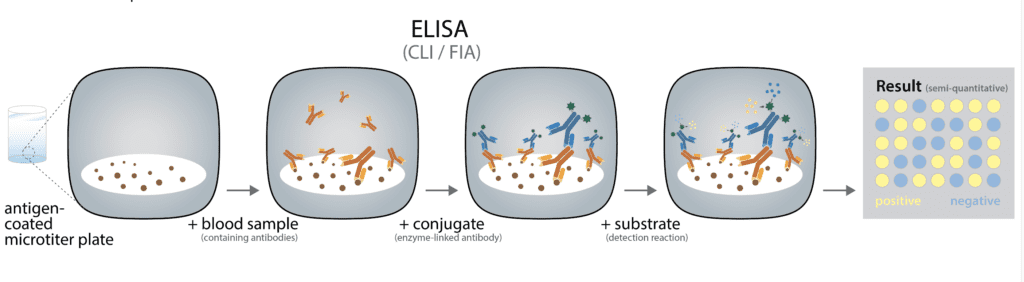
• Enzyme-linked immunosorbent assays (ELISA) offer significantly better specificity and sensitivity compared to RDTs. These tests are generally performed in a laboratory and provide quantitative and qualitative results. Due to the microplate-based design of ELISAs, these tests can be easily automated, which allows for high-throughput screening of hundreds of patient samples at a time. Serological ELISA tests rely on very similar principles as lateral flow tests (Figure 4), most frequently with the roles of the antigen and detecting antibody reversed. The microplate is coated with SARS-CoV-2 antigens, with the antigen of choice most frequently being some variant of the spike protein. The spike protein (S) receptor binding domain (RBD) is thought to be among the most immunogenic epitopes on the viral capsid and is therefore frequently used as an antigen in this test. Another antigen that is highly immunogenic and that plays a major role in T cell responses is the nucleocapsid N. Some ELISA tests use combinations of S and N to increase sensitivity at the cost of a slight reduction in specificity.
The sample, most often blood plasma or serum, is then added to the well, and SARS-CoV-2-specific antibodies contained in the sample will bind to the antigen. An anti-human immunoglobulin antibody conjugated to an enzyme is added, and the antibody-conjugate binds to human antibodies that are bound to the antigen immobilized in the well. Most ELISAs detect either anti-
SARS-CoV-2 IgG, IgM, or IgA, requiring the use of distinct ELISA tests in parallel to detect all three subtypes of immunoglobulins. Wash steps in between every binding step ensure that unbound or lightly unspecific bound components do not interfere. A chromogenic substrate is used to quantify the amount of antibody-conjugate in a color reaction. The strength of the color reaction is directly proportional to the amount of bound antibody. This allows for a semiquantitative interpretation of the antibody levels in a patient’s blood sample, but different binding affinities of the sample antibodies and antibodies contained in the controls impede a fully quantitative analysis. Nonetheless, ELISA tests allow conclusions to be drawn regarding the strength of
the immune system’s reaction in addition to the improved sensitivity and specificity over IST. Also, ELISA tests are used to draw conclusions regarding the seroconversion of each immunoglobulin subclass.
Variants of ELISA are chemiluminescent (CLIA) and fluorescent (FIA) immune assays, where the enzymatic reaction converts a substrate to a chemiluminescent or fluorescent reaction product.
Current Challenges
Scale up of covid-19 diagnostics. Since covid-19 was identified in December 2019, a considerable number of tests have been developed by numerous companies.10 In March 2020, the World Health Organization urged countries to “test, test, test.”11 Widespread testing is critical to map covid-19, monitor its progression rate, and identify hot spots and at-risk populations. However, the number of diagnostic tests currently available on the market does not satisfy the global demand. Many countries are struggling to scale up testing in order to diagnose all symptomatic patients and trace all contacts. In the United States, testing capacity stands at 2.78 tests per thousand people,12 which is 912,396 tests per day. This number is far below the 10 to 30 million tests per day that is estimated to be required to fully reopen the economy while still controlling outbreaks.13,14 The lack of adequate testing prevents early covid-19 detection, which results in continued, unmonitored transmission of the disease.
Rapid antigen detection tests could be a potential alternative solution to molecular testing, due to their suitability for point-of-care settings, short turnaround time, and affordability. However, rapid antigen detection tests show suboptimal sensitivity compared to highly sensitive molecular tests, and for this reason the few antigen tests that have been approved under FDA’s emergency use authorizations (EUA) are recommended to be used only with individuals who are suspected of covid-19 by their healthcare provider within a number of days after the onset of symptoms.15,16 Moreover, the US National Institutes of Health has launched an initiative called Rapid Acceleration of Diagnostics (RADx) to provide a solution to this urgent need for accurate, affordable, easily accessible, and scalable diagnostic testing. RADx has already tasked seven biomedical diagnostic companies to develop a range of new lab-based and point-of-care tests that would improve test availability by millions per week already within the flu season 2020-2021. With national demand estimated to grow rapidly, expectations are high for these new tests to make a significant contribution to monitoring and containing the pandemic.17
European countries are also concerned about testing capacity as they are now facing a second wave of infection. For instance, the UK, which is one of the countries with the highest number of positive cases per day,18 is experiencing a shortage in testing reagents,19 which prevents the effort of a few months ago to scale up the diagnostic testing capacity from 340,000 to 500,000 tests per day by the end of October.20
Better understanding of the immune response to SARS-CoV-2. One of the biggest challenges of this pandemic is to capture the true extent of the virus’s spread and its infection/fatality ratio. Because of the high proportion of asymptomatic or mild infections (approximately 80%), data restricted to laboratory-confirmed cases is not sufficient to provide comprehensive information.21 Therefore, serological detection of specific antibodies against SARS-CoV-2, applied in designed seroprevalence studies, is needed to better estimate the true scale of infections. This approach will provide data on SARS-CoV-2 seroprevalence in different geographical areas or in specific populations such as healthcare workers, pregnant women, or immunosuppressed people. The collected data can then be used by governments to implement public health measures and control strategies and by the private sector to define back-to-work policies.
Seroprevalence studies aim also to understand how the humoral immune response to SARS-CoV-2 infection works and what is the duration of this immunity. Recent studies show that moderate to severe cases of covid-19 will mount a strong humoral immune response, with up to 90% of cases showing robust serum levels of anti-SARS-CoV-2 IgG22-24 with some studies claiming 100% seroconversion rate.25,26
On the other hand, studies looking at asymptomatic patients23 or across a wide range of patients27 show not only a much lower seroconversion rate, but also a correlation of IgG serum positivity and titer with the severity of the disease course. Additionally, asymptomatic cases seem to lose IgG seropositivity faster and more frequently.23 This data suggests that it is still unclear how robust the immune response against an asymptomatic disease manifestation is and whether it will be sufficient to grant immunity against reinfection.
Recently, a virtual workshop on covid-19 serology studies was held to review all the ongoing SARS-CoV-2 serosurvey studies and serological assay performance, as well as identify scientific gaps and develop recommendations for future studies. The conclusion was that although multiple ongoing seroprevalence studies are contributing to a better understanding of the level of SARS-CoV-2 seroconversion in various populations and communities, additional data is still needed to increase our understanding of the immune responses that lead to protection and duration of protection. Such data includes the specific antibody titers that correlate with protection from the disease and viral shedding upon reinfection.26
T cells may also play a critical role in fighting a SARS-CoV-2 infection. It has been shown that patients who have high levels of neutralizing anti-RBD-antibodies are likely to have high levels of nucleocapsid-responsive T cells29 and that SARS-CoV-2-specific CD4+ and CD8+ are frequently found in covid-19 patients.30 Moreover, elevated T cell responses are correlated with recovery,31 and circulating follicular helper T cells reactive to the spike protein are elevated after SARS-CoV-2 exposure and correlate with a patient’s ability to neutralize viral infections.32 Although accumulating evidence supports a role for T cells in covid-19, additional studies are needed to confirm whether they may provide long-term protection from reinfection.
Conclusions
In the past months, we have learned a great deal about this pathogen. The fast spread of the disease and the yet unknown rate of asymptomatic infections, although suggested to be at around 40%, highlight our vulnerability. Early detection of active infections and identification of SARS-CoV-2 seroprevalence combined with public health measures such as social distancing and contact tracing are the best tools that we have to combat the pandemic.
The previously described constraints in diagnostic testing capacity that have emerged suggest that we have to do more to bridge the supply/demand gap. Clinical laboratories can either maximize their existing capacity or establish new capacity. In the first case, clinical labs would need to: compile a full inventory of their installed equipment base, distinguishing between open and closed systems; calculate the maximum theoretical laboratory capacity; and evaluate the potential need for new workflows, additional personnel or alternative suppliers of reagents if open-source systems are used. In the second case, clinical laboratories may consider increasing their equipment footprint by establishing new, high-capacity systems. Closed systems require proprietary reagents and do not provide much flexibility; open systems run a wider range of test methods from multiple suppliers, thereby providing more flexibility and better cost-effective options.

Of course, diagnosis of active infections is not enough to capture the true extent of the viral spread as well as its infection/fatality ratio. More studies need to be done to explain how our immune systems respond to SARS-CoV-2 infection and how long immunity lasts. Financial support from international research agencies and organizations, the coordination of research efforts, and the development of sustained partnerships are essential to providing the best opportunity for controlling this pandemic. l
Albino Troilo, PhD, is a senior marketing manager at Enzo responsible for marketing strategies and marketing communication. He has made major contributions in setting up the molecular diagnostic division at Enzo, which resulted in the launch of several molecular diagnostic products and clinical tests. He is currently leading the global COVID-19 campaign. Troilo holds a PhD in Molecular Health Sciences from ETH Zurich.
References
1. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475-481. doi: 10.1016/S2213-2600(20)30079-5.
2. Kim HS, K JS. Discrepancies between antigen and polymerase chain reaction tests for the detection of rotavirus and norovirus. Ann Clin Lab Sci. 2016;46(3):282-285.
3. Tang YW, Schmitz JE, Persing DH, Stratton CW. The laboratory diagnosis of covid-19 infection: current issues and challenges. J Clin Microbiol. 2020; 58(6):e00512-00520. doi: 10.1128/JCM.00512-20.
4. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1.
5. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. Epub. March 28, 2020. doi:10.1093/cid/ciaa344.
6. Lou B, Li T, Zheng S, et al. Serology characteristics of SARS-CoV-2 infection since exposure and post symptoms onset. Eur Respir J. 2020;56(2):2000763. doi:10.1183/13993003.00763-2020.
7. Long Q-X, Liu B-Z, Deng H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1.
8. Thomas SJ, Nisalak A, Anderson KB, et al. Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: how alterations in assay conditions impact performance. Am J Trop Med Hyg. 2009;81(5):825-833. doi: 10.4269/ajtmh.2009.08-0625.
9. Ratnam S, Gadag V, West R, et al. Comparison of commercial enzyme immunoassay kits with plaque reduction neutralization test for detection of measles virus antibody. J Clin Microbiol. 1995;33(4):811-815. doi: 10.1128/JCM.33.4.811-815.1995.
10. Marca AL, Capuzzo M, Paglia T, et al. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod Biomed Online. 2020;41(3):483-499. doi: 10.1016/j.rbmo.2020.06.001.
11. World Health Organization. WHO Director-General’s opening remarks at the media briefing on COVID-19. March 16, 2020. Available at https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-themedia-briefing-on-covid-19—16-march-2020. Access June 25, 2020.
12. Our World in Data: Coronavirus (COVID-19) Testing. https://ourworldindata.org/coronavirus-testing#how-many-tests-are-performed-each-day. Data at August 19, 2020.
13. Romer PM. Without more tests, America can’t reopen. The Atlantic. April 18, 2020
14. Siddarth D, Weyl GE. Why we must test millions a day. Edmond J. Safra Center for Ethics. April 8, 2020. Available at https://drive.google.com/file/d/1EhUfmT6ayG3ERxX-wZUmB2wtIEOhRAmP/view.
15. Foundation for Innovative New Diagnostics. FIND evaluation update: SARS-CoV-2 immunoassays 2020. https://www.finddx.org/covid-19/sarscov2-eval-immuno. Accessed June 29, 2020.
16. Centers for Medicare & Medicaid Services. https://www.cms.gov/files/document/clia-poc-ag-test-enforcement-discretion.pdf. Accessed October 14, 2020.
17. National Institutes of Health. Rapid Acceleration of Diagnostics (RADX). 2020. https://www.nih.gov/research-training/medical-research-initiatives/radx.
18. European Centre for Disease Prevention and Control. COVID-19 situation update for the EU/EEA and the UK, as of 14 October 2020. https://www.ecdc.europa.eu/en/cases-2019-ncov-eueea. Accessed October 14, 2020.
19. Gross A, Neville S. NHS labs hit by shortage of vital kit and chemicals for covid tests. Financial Times. October 6, 2020 Available at https://www.ft.com/content/aa48893b-9c90-4701-b317-67bda727bbda.
20. Booth R. How prepared is Boris Johnson for a winter resurgence of coronavirus? The Guardian. July 17, 2020. Available at https://www.theguardian.com/world/2020/jul/17/how-prepared-is-boris-johnson-for-a-winter-resurgence-of-coronavirus.
21. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239-1242. doi: 10.1001/jama.2020.2648.
22. Liu T, Wu S, Tao H, et al. Prevalence of IgG antibodies to SARS-CoV-2 in Wuhan – implications for the ability to produce long-lasting protective antibodies against SARS-CoV-2. Preprint. medRxiv. 2020.06.13.20130252. doi: 10.1101/2020.06.13.20130252
23. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200-1204. doi: 10.1038/s41591-020-0965-6.
24. Dogan M, Kozhaya L, Placek L, et al. Novel SARS-CoV-2 specific antibody and neutralization assays reveal wide range of humoral immune response during COVID-19. Preprint. medRxiv. 2020;2020.07.07.20148106. doi: 10.1101/2020.07.07.20148106
25. Suthar MS, Zimmerman M, Kauffman R, et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Med Rep. 2020;1(3):100040. doi: 10.1016/j.xcrm.2020.100040.
26. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845-848. doi:10.1038/s41591-020-0897-1
27. Reifer J, Hayum N, Heszkel B, et al. SARS-CoV-2 IgG antibody responses in New York City. Diagn Microbiol infect Dis. 2020;98(3):115128. doi: 10.1016/j.diagmicrobio.2020.115128.
28. Lerner AM, Eisinger RW, Lowy DR, et al. The covid-19 serology studies workshop: recommendations and challenges. Immunity. 2020;53(1):1-5. doi: 10.1016/j.immuni.2020.06.012.
29. Ni L, Ye F, Cheng ML, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in covid-19 convalescent individuals. Immunity. 2020;52(6):971-977.e3. doi: 10.1016/j.immuni.2020.04.023.
31. Yu HQ, Sun BQ, Fang ZF, et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur Respir J. Epub. August 27, 2020. 2020;56(2):2001526. doi: 10.1183/13993003.01526-2020.
31. Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020;101(8):791-797. doi: 1099/jgv.0.001439.
32. Weiskopf D, Schmitz KS, Raadsen MP, et al. Phenotype of SARS-CoV-2-specific T-cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48): eabd2071. doi: 10.1126/sciimmunol.abd2071.
Featured image: Colorized scanning electron micrograph of a VERO E6 cell (blue) heavily infected with SARS-COV-2 virus particles (orange), isolated from a patient sample. Image captured and color-enhanced at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID





Great article!
The author has failed to mention that Covid-19 antibody tests are also saliva based. See below
https://covabscreen.com/training/
Also IgA is considered an important neutralizing antibody in the mucosal area.