With the Omicron variant becoming the dominant mutation of SARS-CoV-2, it is critical to understand how this variant works to find ways to diagnose and combat it and future variant types.
By GARETH HUMPHRIES
In November 2021, a new variant of concern (VoC) was designated Omicron. Having first been identified in South Africa, Omicron has quickly displaced Delta to become the dominant variant throughout much of the world.
In addition to increased transmissibility, data suggests that Omicron has the capacity for substantial immune evasion, threatening to undermine the population immunity conferred by vaccination programs.
Omicron Emergence and Mutations
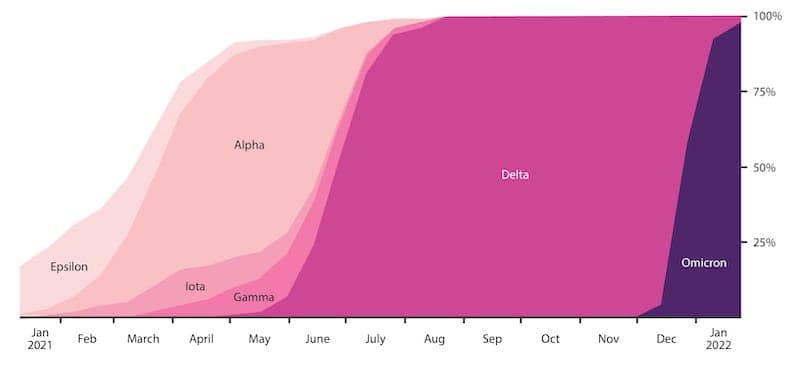
In November 2021, a new variant was added to the SARS-CoV-2 family. Rapidly outcompeting the incumbent Delta in South Africa, the new variant rose from a test positivity rate of less than 1% in late October, to 35% by the end of the following month1. On November 26, the virus was classified as lineage B.1.1.529 and designated Omicron by the World Health Organization, making it the fifth VoC2. By the end of 2021, Omicron had been detected in over 90 countries and has now become the predominant form of SARS-CoV-2 (Figure 1), coinciding with a new global wave of infections.
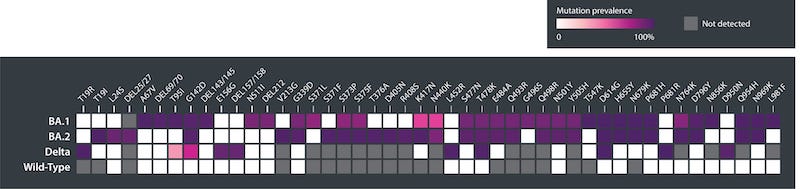
What has made Omicron an outlier from other variants is the degree to which it has mutated. Compared to wild-type SARS-CoV-2, Omicron has 50 to 60 mutations (Figure 2). Around 30 of these are found in its Spike gene, roughly twice the number found in the Delta variant5. Fifteen of the 30 Spike mutations identified are consistently found in Omicron’s receptor-binding domain (RBD), the immunodominant region responsible for ACE2 binding and the major target of neutralizing antibodies. Of these, G339D, N440K, S477N, T478K, Q498R and N501Y have already shown to enhance ACE2 binding in previous SARS-CoV-2 variants1,6,7 . Most of the remaining mutations have only been found in rare isolates, making their appearance in Omicron all the more surprising.
Omicron Variant’s Effect on Diagnostics
As the Omicron variant spread in South Africa in late 2021, its emergence coincided with an increasing frequency of Spike gene target failure (SGTF) in RT-PCR assays1 —a form of diagnostic failure in which primers designed against mutation hotspots are unable to bind, resulting in no target amplification. Like the Alpha variant, the Omicron variant contains a 69–70 deletion in its Spike protein that causes SGTF in certain types of assays, of which incurred 90% SGTF by the end of November 2021 in South Africa1. However, as these multiplex assays are still able to yield true positives via other target genes, SGTF has made for a good surrogate of variant detection9. Prevalence and spread of Omicron has, therefore, been closely tracked through a combination SGTF and sequencing to support policy making and the distribution of resources10.
Unsurprisingly, serological tests for SARS-CoV-2 are significantly affected by Omicron. Most rapid tests use nucleoprotein (NP) as a target, given its high immunogenicity and exclusion from most vaccine formulations. Omicron’s NP includes the P13L, R203K, and G204R substitutions, as well as the Δ31–33 deletion, resulting in reduced binding affinity, and thus sensitivity. A study, published in early January, found that rapid antigen tests were delayed in their ability to detect COVID-19 during early symptoms onset11, and following studies have shown considerable heterogeneity in test sensitivity12. Combined with the shorter timeframe from exposure to infectiousness, this has undermined the accuracy of lateral flow testing and, again, put into question its role in disease diagnosis.
New Phenotypes
The combination of the Omicron variant’s Spike mutations is estimated to double (x2.4) the affinity of its RBD for ACE2 in comparison to the Wuhan Hu-1 wild-type1. It is partly for this reason that Omicron is more transmissible than previous VoCs. However, there are a multitude of other factors that sets Omicron apart.
A hallmark of the SARS-CoV-2 Spike protein is its furin cleavage site, located between its S1 and S2 subunits. Cleavage at this site increases flexibility and liberates key binding regions to facilitate Spike’s binding to ACE2 and entry to the host cell13. The major protease involved in this reaction is TMPRSS2—a human cell-surface protein whose natural biological function is largely unknown14. TMPRS22 shows greater distribution in the lower respiratory tract, resulting in SARS-CoV-2’s tropism for this region and some of the tell-tale symptoms of COVID-19, such as coughing, breathing difficulties, and pneumonia15.
Bucking the trend, the Omicron variant’s Spike shows reduced cleavage efficiency compared to other variants, and has, in fact, shown to be impaired in mediating cell entry in the presence of TMRPRSS216. At first, the findings appeared to contradict Omicron’s high rate of reproduction and transmission. However, studies have since shown that this is compensated for by Omicron’s improved use of the ubiquitous endosomal pathway of cell entry as opposed to cell-surface fusion17,18 .
Because of Omicron’s decreased reliance on TMPRSS2-dependent cell entry, its host tropisms are also altered, with improved ability to infect and replicate in bronchial cells and nasal cavities, compared with the alveoli of the lungs19,20 . As a result, it is thought that Omicron is more readily excreted to the oral-nasal cavities where it can spread easily. For this same reason, Omicron is thought to result in reduced disease severity, as the tropisms associated with more severe forms of COVID-19 are diminished.
Omicron Variant Immunity: The Great Escape?
While much remains to be understood about the Omicron variant, what’s been clear from the outset is its ability to evade vaccine- and natural-induced immunity. For recipients of major vaccines, studies have found low-to-undetectable levels of neutralizing antibodies against Omicron antigens21. The majority patient-derived monoclonal antibodies show no binding, and neutralizing activity of polyclonal sera have shown to be 10- to 25-fold less effective than other variants5,22,23. As a result, antibody immunity against Omicron is only partial, with an estimated potential breakthrough rate of over 70%1. Work to define the specific mutations that facilitate immune escape is still underway. However, it is clear that seven of Omicron’s RBD mutations significantly impact RBD-neutralizing therapeutic antibodies. Coupled with substitutions, deletions, and an insertion in the N-terminal domain, these mutations are likely to underlie the reduced sensitivity of antibodies to Omicron1.
Consistent with previous findings, CD4+ and CD8+ T-cells elicited from vaccination show extensive cross-reactivity with Omicron (70–80%) and are expected to play a dominant role in protecting from severe disease24,25 . This is explained by the ability of T-cells to recognize a broader range of more highly conserved Spike epitopes that are less susceptible to mutation. Moreover, while the longevity of cell-mediated immunity isn’t fully known, survivors of the original SARS-CoV virus have shown robust T-cell responses 17 years post-infection, suggesting durable protection26. Combined with Omicron’s attenuated virulence, the resilience of cellular immunity explains the divergence between new and severe cases of COVID-19 in recent months.
Omicron Variant Sub-Lineage BA.2
Shortly after Omicron was identified, distinct sub-lineages of the original BA.1 variant were identified, now including BA.1.1, BA.2 and BA.327. Of these, BA.2 has gained the most attention, following its rapid proliferation in Europe and Asia. Outcompeting the BA.1, BA.2 is around 1.5 times more transmissible and is soon becoming the new dominant form of SARS-CoV-2 globally28.
Of the 60 or so Omicron mutations, only around half are shared between BA.1 and BA.2, while the remaining 30 are unique to the BA.2 sub-lineage—comparable to the genetic differences of Omicron and Delta. Because of this, BA.2 is serologically distinct from BA.1, with cases of BA.2 infection having been reported in BA.1-recovered patients29,30 .
Complicating detection, BA.2 does not contain the 69–70 deletion in its Spike gene, and so cannot be detected by SGTF—hence the “stealth variant” moniker. In regard to pathogenesis, BA.2 shows better replication efficiency in nasal cells, as well as increased virulence in animal models30. However, findings on disease in humans have so far been inconclusive, with no significant differences apparent for patient outcomes31.
The reason for BA.2’s rapid ascent isn’t yet clear. An early hypothesis was that BA.2’s Spike mutations allow it to evade a degree of BA.1 and Delta immunity, giving it an edge over these, and other variants. However, recent data appear to refute this, with studies showing good cross-protection between Omicron sub-lineages32. Therefore, further work is needed to understand what is driving BA.2, to contain its spread and prevent the emergence of similar variants.
Future Perspectives
With the Omicron variant’s reach being so comprehensive, there is reason to hope that the recent wave could be the pandemic’s last. The continued attenuation of SARS-CoV-2 into its current form has uncoupled caseloads and incidence of severe disease, while boosting population immunity. Yet, due to the diversity in exposure to different vaccines and variants, population immunity has become highly heterogenous, with individuals displaying markedly different abilities to fight off infection. Therefore, considerable research is still needed to determine the most at-risk groups for prioritizing boosters. Omicron is just the latest in a string of variants to exhibit high antigenic and functional plasticity, meaning further shifts in serology and disease should be anticipated.
The emergence of Omicron has highlighted how fragile antibody-mediated protection can be, undermining correlates of protection for both infection- and vaccine-induced immunity. Further work is, therefore, needed to profile cellular correlates and develop improved means of interrogating this form of immunity at population-scale.
About the Author
Gareth Humphries is the business development manager for the Native Antigen company. He graduated with a degree in chemistry from the University of Bath, and worked in drug discovery for some years before stepping into the “suit & tie” side of science. Working for globally recognized distributors VWR International and later Thermo Fisher, he was a sales specialist serving the Oxfordshire life-science community for over 10 years. Humphries joined The Native Antigen Company at the start of 2019; supporting the company’s global customer base with reagents for virology, toxicology, and more recently, a broad range of human coronavirus products, which have found extensive use globally in the response to the SARS-CoV2 pandemic.
References
1. Viana, R., Moyo, S., Amoako, DG., Tegally, H., Scheepers, C., Althaus, CL., Anyaneji, UJ., Bester, PA., Boni, MF., Chand, M., Choga, WT. Colquhoun, R., Davids, M., Deforche, K., Doolabh, D., Plessis, LD., Engelbrecht, S., Everatt, J., Giandhari, J., Giovanetti, M… Oliveira, TD. (2022). Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. https://doi.org/10.1038/s41586-022-04411-y
2. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. (2020). World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern (who.int)
3. Corum, J. and Zimmer, C. (2022). Tracking Omicron and Other Coronavirus Variants. The New York Times. https://www.nytimes.com/interactive/2021/health/coronavirus-variant-tracker.html
4. Overview of Variants in Countries. CoVariants. https://covariants.org/per-country
5. VanBlargan, LA., Errico, JM., Halfmann, PJ., Zost, SJ., Crowe, JE., Purcell, LA., Kawaoka, Y., Corti, D., Fremont, DH., Diamond, MS. (2022). An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nature Medicine. https://doi.org/10.1038/s41591-021-01678-y
6. Cameroni, E., Bowen, JE., Rosen, LE., Saliba, C., Zepeda, SK., Culap, K., Pinto, D., VanBlargan, LA., Marco, AD., Lulio, JD., Zatta, F., Kaiser, H., Noack, J., Farhat, N., Czudnochowskie, N., Havenar-Daughton, C., Sprouse, KR., Dillen, JR., Powell, AE., Chen, A… Corti, D. (2022). Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602, 664-670. https://doi.org/10.1038/s41586-021-04386-2
7. Woo, HG., Shah, M. (2022). Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front. Immunol. https://doi.org/10.3389/fimmu.2021.830527
8. SARS-CoV-12 (hCoV-19) Mutation Reports: Lineage Comparison. Outbreak Info. https://outbreak.info/compare-lineages?pango=BA.1&pango=BA.2&gene=ORF1a&gene=ORF1b&gene=S&gene=ORF3a&gene=E&gene=M&gene=ORF6&gene=ORF7a&gene=ORF7b&gene=ORF8&gene=N&gene=ORF10&threshold=75&nthresh=1&sub=false&dark=true
9. Brown, KA., Gubbay, J., Hopkins, J., Patel, S., Buchan, SA. Daneman, N., Goneau, LW. (2021). JAMA. 2021;325(20):2115-2116. https://doi.org/10.1001/jama.2021.5607
10. Ferguson, N., Ghani, A., Cori, A., Hogan, A., Hinsley, W., Volz., E. (2021). Report 49 – Growth, population distribution and immune escape of Omicron in England. Imperial College London. https://writix.co.uk/blog/covid-19-report-49-omicron
11. Adamson B., Sikka R., Wyllie A. L., Premsrirut. P. Discordant SARS-CoV-2 PCR and Rapid Antigen Test Results When Infectious: A December 2021 Occupational Case Series. (2022) medRxiv; https://doi.org/10.1101/2022.01.04.22268770
12. Bekliz M. et al. Sensitivity of SARS-CoV-2 antigen-detecting rapid tests for Omicron variant.(2021) medRxiv; https://doi.org/10.1101/2021.12.18.21268018
13. Peacock, T.P., Goldhill, D.H., Zhou, J. et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 6, 899–909 (2021). https://doi.org/10.1038/s41564-021-00908-w
14. Rahman, N., Basharat, Z., Yousuf, M., Castaldo, G., Rastrelli, L., & Khan, H. (2020). Virtual Screening of Natural Products against Type II Transmembrane Serine Protease (TMPRSS2), the Priming Agent of Coronavirus 2 (SARS-CoV-2). Molecules (Basel, Switzerland), 25(10), 2271. https://doi.org/10.3390/molecules25102271
15. Murphy K. (2020). SARS CoV-2 Detection From Upper and Lower Respiratory Tract Specimens: Diagnostic and Infection Control Implications. Chest, 158(5), 1804–1805. https://doi.org/10.1016/j.chest.2020.07.061
16. Meng, B., Abdullahi, A., Ferreira, I.A.T.M. et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature (2022). https://doi.org/10.1038/s41586-022-04474-x
17. Hui, K. P. Y. et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. (2022). Nature https://doi.org/10.1038/s41586-022-04479-6
18. Willet. B et al. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv (2022). https://doi.org/10.1101/2022.01.03.21268111
19. HKUMed finds Omicron SARS-CoV-2 can infect faster and better than Delta in human bronchus but with less severe infection in lung. The University of Hong Kong. https://www.med.hku.hk/en/news/press/20211215-omicron-sars-cov-2-infection
20. Peacock T. et al. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv (2021). https://doi.org/10.1101/2021.12.31.474653
21. Flemming, A. Omicron, the great escape artist. Nat Rev Immunol 22, 75 (2022). https://doi.org/10.1038/s41577-022-00676-6
22. Cheng, S.M.S., Mok, C.K.P., Leung, Y.W.Y. et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med (2022). https://doi.org/10.1038/s41591-022-01704-7
23. Dejnirattisai W. et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 183,3 (2022) https://doi.org/10.1016/j.cell.2021.12.046
24. Liu, J., Chandrashekar, A., Sellers, D. et al. Vaccines Elicit Highly Conserved Cellular Immunity to SARS-CoV-2 Omicron. Nature (2022). https://doi.org/10.1038/s41586-022-04465-y
25. Keeton, R., Tincho, M.B., Ngomti, A. et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature (2022). https://doi.org/10.1038/s41586-022-04460-3
26. Le Bert, N., Tan, A.T., Kunasegaran, K. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462 (2020). https://doi.org/10.1038/s41586-020-2550-z
27. Kumar, S., Karuppanan, K. and Subramaniam. G. (2022). Omicron (BA.1) and Sub-Variants (BA.1, BA.2 and BA.3) of SARS-CoV-2 Spike Infectivity and Pathogenicity: A Comparative Sequence and Structural-based Computational Assessment. BioRxiv https://doi.org/10.1101/2022.02.11.480029
28. Zhou, H., Tada, T., Dcosta, BM. And Landau, NR. (2020). Neutralization of SARS-CoV-2 Omicron BA.2 by Therapeutic Monoclonal Antibodies. BioRxiv https://doi.org/10.1101/2022.02.15.480166
29. Chen, J. and Wei, GW. (2022). Omicron BA.2 (B.1.1.529.2): high potential to becoming the next dominating variant. Cornell University https://doi.org/10.48550/arXiv.2202.05031
30. Yamasoba, D. et al. (2022). Virological characteristics of SARS-CoV-2 BA.2 variant. BioRxiv https://doi.org/10.1101/2022.02.14.480335
31. Wolter, N., Jassat, W., Gottberg, AV. And Cohen, C. (2022). Clinical severity of Omicron sub-lineage BA.2 compared to BA.1 in South Africa. MedRxiv https://doi.org/10.1101/2022.02.17.22271030
32. Chemaitelly, H. et al. (2022). Protection of Omicron sub-lineage infection against reinfection with another Omicron sub-lineage. MedRxiv https://doi.org/10.1101/2022.02.24.22271440