Qlucore, Lund, Sweden, has released Qlucore Omics Explorer (QOE) version 3.7, an extensive upgrade adding features in many different areas to strengthen visual analysis support even further.
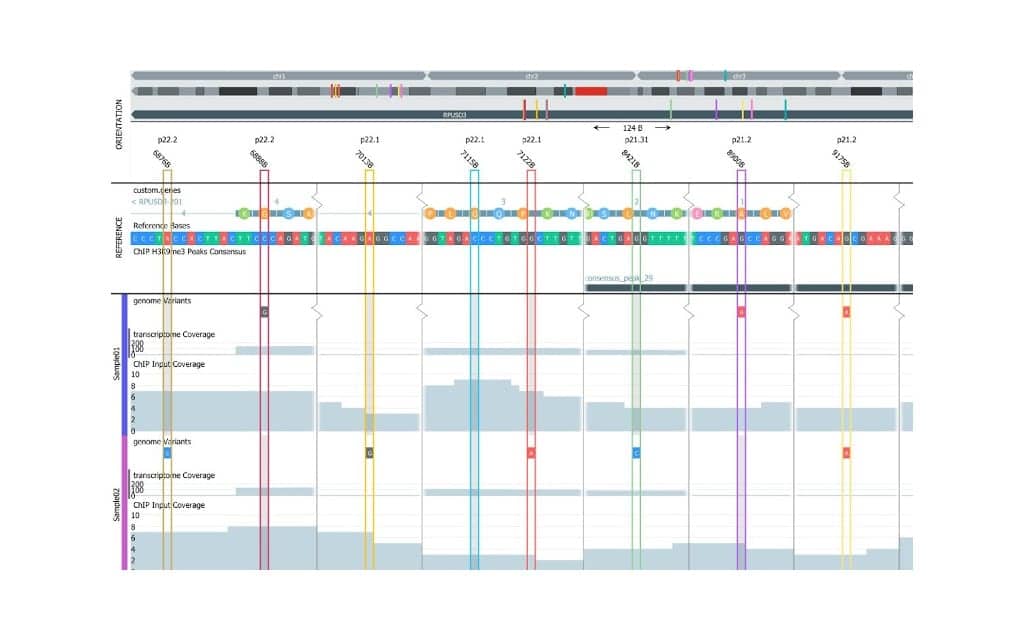
Key features of the new version are ChIP-seq and ATAC-seq analysis support, a new biomarker workbench coupled with an upgraded statistical framework, and significant enhancements to the genome browser. The genome browser has new visualization functions plus support for ChIP-seq, ATAC-seq, and gene fusions. The biomarker workbench is optimized for experiments and studies in the areas of drug development and biomarker discovery.
Other new features include:
- The possibility to add so-called restrictions when setting up statistical tests.
- Easy support for Two-way ANOVA, test the combination effect of two sample annotations.
- A new 2D plot, pie chart – making it possible to visualize the distribution of samples.
- Box plots updates, now possible to display a group of variables for each box.
- New color palette, including option to select color based on the annotation group name.
“We’re thrilled to announce the latest version of the Qlucore software which features a lightning-fast genome browser with best-in-class support for viewing and navigating gene fusions,” says Qlucore Founder and President Carl-Johan Ivarsson. “Qlucore software now enables even faster visualization of scientific data experiments and means researchers no longer have to depend on an expert in bioinformatics to explore and analyze Omics and NGS data sets.”
For more information, visit Qlucore.
Featured image: The new Qlucore Omics Explorer release (3.7) is an extensive upgrade, featuring an improved genome browser, pictured. (Courtesy: Qlucore)