Genomenon Inc, Ann Arbor, Mich, has announced a partnership with Google, Menlo Park, Calif, to make the company’s genomic mutation data available on the Google Cloud platform. Under the agreement, Genomenon’s cited variants reference (CVR) database will be made publicly available for use in genomic applications via BigQuery, Google Cloud’s big data and machine learning data warehouse.
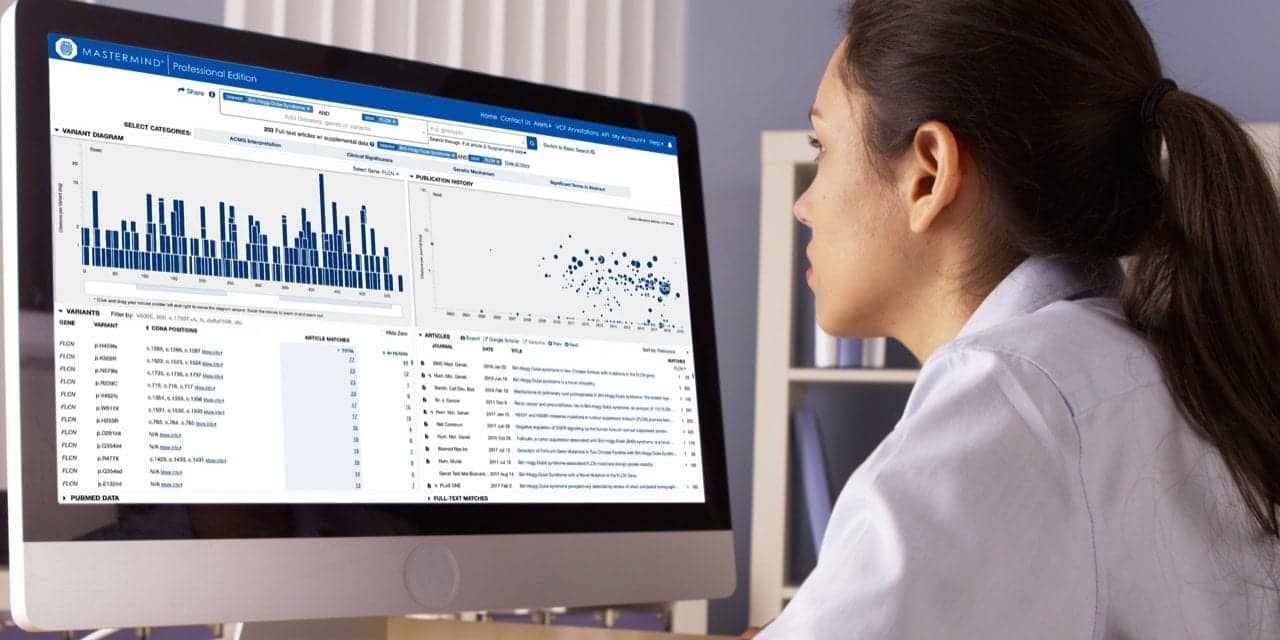
Genomenon powers evidence-based genomics in support of faster and more comprehensive diagnostic and treatment decisions. The company’s flagship product, the Mastermind genomic search engine, provides immediate insight into the published genomic research about every disease, gene, and variant found in the literature. The search engine scans the titles and abstracts of more than 30 million scientific and medical publications listed in the National Library of Medicine’s PubMed database. For articles found to contain genomic information, the full text is then indexed to develop a comprehensive view of the genomic literature. To date, the Mastermind genomic search engine has indexed the text of more than 6.2 million genomic publications and identified more than 4.1 million references to genomic variants.
Used by hundreds of diagnostic labs around the world, Mastermind accelerates genomic interpretation by providing unique insight into genomic relationships found in the full text of millions of scientific articles. Pharmaceutical researchers also license the database in order to identify and prioritize genomic biomarkers for drug discovery and clinical trial targets.
Generated using the Mastermind genomic search engine, Genomenon’s CVR database is one of the most comprehensive indexes of genomic literature in the world, encompassing more than 4.1 million references to genomic variants found in medical literature. Each variant is annotated with a citation count based on the number of scientific publications mentioning the variant, along with a link to the Mastermind genomic search engine to view full search results for those articles.
The CVR database is designed to help clinicians and researchers prioritize and scale their genomic interpretation. Because it is based on the presence of evidence in the literature, the CVR database can be used in genomic analysis pipelines as an evidence filter for clinical actionability, and as a quick way to gain insights into the literature for variant curation. The CVR database is also useful for researchers exploring novel, unpublished variants across patient cohort genomic datasets, by enabling them to search for variants with little or no evidence in the medical literature.
For further information, visit Genomenon.