Summary:
Researchers at Shandong University have developed a CRISPR-Cas10-integrated graphene biosensor capable of amplification-free, ultra-sensitive RNA and miRNA detection at attomolar levels, offering a powerful tool for molecular diagnostics.
Takeaways:
- Unmatched Sensitivity: The platform detects RNAs and miRNAs at incredibly low concentrations (as low as 214 aM) without the need for amplification.
- Robust, Label-Free Design: The system leverages continuous CRISPR-Cas10 activity and graphene transistors for rapid, label-free, and specific detection.
- Scalable for Real-World Use: With its simple, cost-effective design, the biosensor is well-suited for point-of-care diagnostics and future integration into portable health monitoring devices.
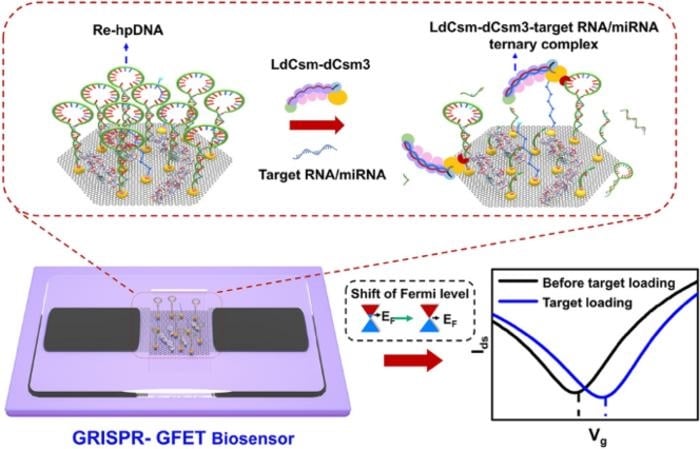
Researchers from Shandong University have developed a novel biosensing platform that integrates the CRISPR-Cas10 system with graphene field-effect transistors (GFETs) for amplification-free, highly sensitive, and specific RNA and microRNA (miRNA) detection. This innovative approach achieves detection limits at the attomolar (aM) level, making it a powerful tool for health monitoring and disease diagnosis.
Why the CRISPR-Cas10-GFET Biosensor Matters
- High Sensitivity: The biosensor demonstrates exceptional sensitivity, with detection limits as low as 214 aM for medium-length RNAs and 427 aM for miRNAs.
- Amplification-Free Detection: By leveraging the continuous ssDNA cleavage activity of the CRISPR-Cas10 system and the high charge density of hairpin DNA reporters on the GFET channel, the biosensor achieves label-free and amplification-free detection.
- Versatility: The platform is capable of detecting a wide range of RNA and miRNA targets, including clinically relevant biomarkers such as miRNA-155 for breast cancer detection.
Innovative Design and Mechanisms
- CRISPR-Cas10 System: The type III CRISPR-Cas10 system provides a unique mechanism for RNA detection through the continuous cleavage of ssDNA reporters activated by target RNA. A mutation in the Csm3 subunit (Csm3D34A) prevents the degradation of target RNA, allowing sustained cleavage activity.
- Graphene Field-Effect Transistors (GFETs): The GFETs offer high sensitivity and rapid response capabilities due to the unique electronic properties of graphene. The high charge density of hairpin DNA reporters on the GFET channel enhances the detection signal.
- Integration: The combination of CRISPR-Cas10 with GFETs creates a synergistic platform that leverages the strengths of both technologies for highly sensitive and specific nucleic acid detection.
Future Outlook
- Scalability and Practical Applications: The CRISPR-GFET biosensor is scalable and suitable for point-of-care testing due to its simplicity, low cost, and rapid detection capabilities. It mitigates the risks associated with nucleic acid amplification and cross-contamination, making it a versatile diagnostic tool.
- Further Research: Future work may focus on optimizing the biosensor for multiplex detection of multiple RNA targets and integrating it with portable devices for real-time monitoring and remote diagnostics.
- Mechanistic Insights: This study provides valuable insights into the mechanisms underlying the CRISPR-Cas10 system and its integration with GFETs, offering a promising path for the development of advanced nucleic acid detection technologies.
Stay tuned for more groundbreaking advancements from the research team at Shandong University as they continue to explore innovative solutions for nucleic acid detection and molecular diagnostics.
Featured Image: A collaborative biosensing system is developed for amplification-free RNA/miRNA detection by integrating Type III CRISPR-Cas10 system with a graphene field-effect transistor (GFET). Continuous cleavage of ssDNA by the mutant CRISPR-Cas10 effector complexes and high charge density of hairpin DNA reporters on the GFET channel enable the detection limit to reach the level of aM. A universal sensing detection platform is established to directly detect the medium-length RNAs and miRNAs in clinical samples with the recognition capability of single nucleic acid. Image: Mingyuan Sun, Zhenxiao Yu, Shuai Wang, Jiaoyan Qiu, Yuzhen Huang, Xiaoshuang Chen, Yunhong Zhang, Chao Wang, Xue Zhang, Yanbo Liang, Hong Liu, Qunxin She, Yu Zhang, Lin Han.