You are standing in an oncology waiting room with three other cancer patients. You have each been given a prescription for an oncology or chemotherapeutic drug. You turn over the thought in your mind … will I get a wig, or just wear a hat? You slowly read the awful list of side effects provided to you by the nurse. You may feel tired, vomit constantly, experience muscle weakness, alopecia, suffer major organ damage, and other severe side effects. Well, you think, at least this therapy will give me a fighting chance to be well again. Then, the nurse announces that while each of you will likely experience negative side effects, on average only one of the four of you will actually be helped by the drug you are sacrificing so much to take.
Unfortunately, for many this is an authentic scenario. Improving drug effectiveness is one of the major factors driving the adoption of companion diagnostics tests for oncology therapeutics. A study by the Tufts University Center for the Study of Drug Development, Boston, demonstrates that, for oncology therapeutics as a class, only 25% of patients taking these compounds have a positive clinical response. This compares with an average response rate of 50% for medications as a whole.
Clearly, a better paradigm is needed. One significant part of the solution has been the trend toward laboratory tests that predict which patients will respond to a given drug. These tests may identify subpopulations of patients who have genetic or proteomic profiles that will cause them to respond to a medication, or who may be at increased risk of serious side effects, effectively personalizing or individualizing drug therapy. These unique tests are called companion diagnostics (formerly theranostics), and constitute a large component of how personalized medicine will be translated into the laboratory.

Mark Cross
is the senior director of sales and marketing for Biocare Medical LLC, Concord, Calif.
Kent Bottles, MD, senior fellow at the Thomas Jefferson University School of Population Health, Philadelphia, writes that personalized medicine is a trend that health care executives can’t ignore.1 Personalized medicine seeks to translate genomic and follow on proteomic discoveries into effective therapies based on each individual’s genetic makeup. Companion diagnostics (CDx) is a major driver of personalized medicine, bringing with it the potential to expand the leadership role of the laboratory and pathology community.
A personalized medicine approach concentrates more on the unique characteristics of each individual as opposed to a population. “Advances in genomics and digital technology are making it possible to shift the focus of evidence-based medicine from the population to the individual patient,” Bottles writes. “Today, drug treatment and disease screening follow a one-size-fits-all approach that leads to overtreatment and unnecessary expense. Genetic testing allows us to individualize the treatment for the patients.”1
Companion diagnostics gives an entirely new viewpoint on the statistic that only one in four patients respond to the average cancer medication. From a CDx viewpoint, drugs that are not effective in an individual patient are not necessarily ineffective; they are simply not the right drug for that particular patient. For modern drugs in which the drug target and mechanism as well as disease biology are well understood, this is an issue that development of an appropriate CDx test may be able to resolve.
An example of the effectiveness of a CDx test is determination of a non-small-cell lung cancer (NSCLC) patient’s EGFR gene mutation status in order to predict treatment response to erlotinib (Tarceva®) or gefitinib (Iressa®). Both of these therapeutics offer better outcome on tumors that have mutant EGFR genes that cause the tumor to receive an abnormal level of growth signals through the EGFR signaling pathway. Patients who are shown to be nonresponders can be put on alternative therapies instead of wasting valuable time on therapies that will not treat their disease, and also avoid the significant toxic side effects of these medications.
CDx tests are unique because they require sophisticated, multimillion-dollar clinical trials similar to those required for drugs. These clinical trials are designed to prove that the CDx test accurately separates patients who will respond to a given medication from likely nonresponders. In this article, I will focus on one of the most rapid adopters of the companion diagnostic paradigm: oncology and the anatomic pathology laboratory.

Table 1
WHAT IS DRIVING CDX?
If all pharmaceutical therapies worked equally well in all patients, there would be no need for companion diagnostic testing. Different patients can have radically different responses to certain drug therapies, driven by their innate genetic, epigenetic, and proteomic diversity. This is exacerbated for drugs in oncology since each tumor adds additional heterogeneity, with many subject to multiple mutations, arising from the diversity of the random mutational process that gives rise to cancer cells and the resultant genetic instability.
Companion diagnostics brings a counterweight to this huge pool of genetic diversity. These tests allow therapeutic strategies to be developed based on the unique characteristics of each patient and their disease profile, so that they are custom fitted like a tailored suit or a knee replacement.
Further, the urgency involved, especially in late-stage cancer patients, does not allow clinicians or patients the luxury of sequentially using several medications to find out which one will be most effective. First, the disease may spread over time. And second, survival is an issue for some diseases: taking a trial-and-error approach on a lung cancer patient results in more than 42% of these patients losing their lives within the first year.2
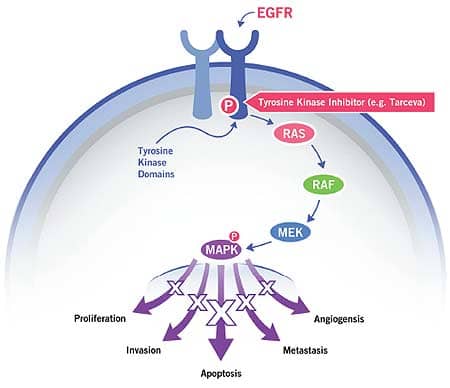
Figure 1: A mutant, constitutively active EGFR is inhibited by Tarceva, a drug that turns off growth signals in some NSCLC tumors.
HOW CDX TESTS WORK
Many advanced oncology medications target specific tumor types by binding to mutant genes or the proteins they express. This may inhibit oncogenic functions that support tumor development or amplify signaling pathways that suppress cancer development, such as:
- Unusually rapid cell growth, proliferation, protein synthesis, or metabolism;
- Increased vascularization to increase the tumor’s nutrient supply;
- A breakdown in cell junctions that may foster metastasis; and
- The natural apoptotic (cell death) response when chromosome instability, mutations, or inappropriate protein activity are detected.
The EGFR pathway in Figure 1 illustrates how a signal from outside a cell can affect cellular function. In this case, epidermal growth factor (EGF) binds to the EGF receptor (EGFR), resulting in a conformational change in the protein. This shape change in EGFR results in a binding site in the tyrosine kinase domain being exposed. It is then able to bind and be energized (phosphorylated) by ATP (adenosine 5′-triphosphate). Chemical signals are passed from the phosphorylated tyrosine kinase domain to the RAS protein, then on to RAF, MEK, and MAPK. MAPK signals enter the nucleus, affecting levels of cell proliferation, invasion, apoptosis, metastasis, and angiogenesis.
Normally, these events are controlled very carefully by the cell with the EGFR signaling pathway dialed up or down as needed. However, in some cancers, an EGFR mutation causes dysregulation of this signaling pathway, resulting in pathogenesis and progression of cancer. The tyrosine kinase domain of the protein becomes constitutively active, wherein the growth signals are permanently turned on. This can lead to uncontrolled cell growth; increased angiogenesis (blood vessel formation), which provides more nutrients to tumors; the ability to metastasize; and failure of natural apoptotic pathways that are designed to cause cell death in the event that mutations or aberrant protein activity is sensed by the cell. All of these conditions favor survival of cancer cells.
How can an inhibitor molecule attenuate these growth signals? In Figure 1, an EGFR inhibitor called Tarceva (erlotinib) binds to the now permanently exposed tyrosine kinase domain binding site in mutant EGFR. However, it does not produce the signals that will activate subsequent proteins in the signaling pathway. Therefore, inappropriate cell proliferation, metastasis, and angiogenesis is attenuated and appropriate apoptosis is restored.
Another example is the breast cancer drug Herceptin® (trastuzumab), a therapeutic antibody that operates by binding to and inhibiting the HER2 receptor, which mediates cell growth. In some patients, the HER2 gene is amplified, with some cancer cells showing five, 10, 20, or more copies of this gene. Since HER2 regulates cell growth, these cells receive an abnormally high level of growth signals, which can cause the tumor cells to proliferate rapidly, resulting in a poor prognosis. Herceptin blocks these receptors, which may result in reduction of the cancer growth rate, and also flags the tumor cell for destruction by the immune system.
CDx tests for HER2 allow use of either DNA probes to bind to individual HER2 genes, or antibodies to mark the HER2 (c-erbB-2) protein receptors in the cell membrane. The pathologist then uses a specific algorithm3 to determine if the HER2 gene or receptor is amplified. For patients with amplified HER2, the cancer is likely to respond to Herceptin. If the gene or receptor is not amplified, then that tumor does not have the cell-overgrowth problem that Herceptin is designed to attenuate, and that patient’s tumor is not likely to respond to Herceptin therapy.
Tests to Aid in Differentiation of Lung SqCC Versus Lung Adenocarcinoma
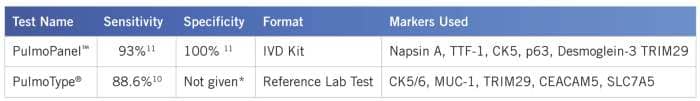
Table 2: PulmoType® and PulmoPanel™ were developed by Clarient and Biocare Medical, respectively.*Pulmotype specificity data not published, but reported as similar to that of combined p63 and TTF-1.10
CRITICAL DIAGNOSES NECESSARY FOR DRUG EFFICACY
In some cases, no companion diagnostic test is referred to in the drug labeling, but it is stated that the therapy should only be used for patients with very specific disease states. One example of this is in NSCLC. Historically, once it was determined that a patient had NSCLC, there was no need for any further differentiation of the disease, as the treatment was the same for all NSCLC tumors. In recent years, it has been demonstrated that certain of the new targeted drugs can cause serious side effects or have reduced efficacy if given to patients with the wrong NSCLC histology subtype. For example, the drug Avastin® inhibits the protein VEGF, which functions to increase vascularization in tumors, promoting growth. However, Avastin has been shown to cause pulmonary hemorrhage, resulting in a 30% fatality rate in a common type of NSCLC called squamous cell carcinoma (SqCC).4 Also, pemetrexed, a folate analog, should be prescribed for lung adenocarcinoma, but not for SqCC.
Further, about 70% of NSCLC diagnoses are made based on a small biopsy or fine needle aspirate sample since the lung cancer tissue is often not resected.5 This presents considerable diagnostic challenges, and even experts may not be consistent in determining histotype.6 In a study comparing diagnostic accuracy on preoperative bronchial biopsy specimens and the subsequent resected lung tissue, diagnostic accuracy has been shown to be 75% for SqCC. Diagnosis of adenocarcinoma was more challenging, resulting in a diagnostic accuracy of 50%.7
So, while there is no regulatory requirement for a companion diagnostic test, an ancillary test that would aid in differentiation of squamous cell carcinoma versus adenocarcinoma in NSCLC with high sensitivity and specificity is critical. Until recently, percent specificity and sensitivity for tests of this type were in the range of the 70s to the low 80s.8 Clearly, there was a need for tests with a higher level of accuracy to avoid the startling mortality rate in SqCC patients. More recently, some tests have been made available that provide greatly increased sensitivity and specificity, giving the treating physician greater confidence in prescribing these medications.9,10,11
Novel tools to aid in differentiation of lung adeno- versus SqCC (Table 2) have recently been developed. These feature greatly increased sensitivity (Pulmotype and PulmoPanel) and specificity (PulmoPanel) compared to routinely used methods. Both methods have very low equivocal rates—11% for Pulmotype10 and 7% for PulmoPanel11 compared to 22% for the more traditional p63 and TTF-1.10 This may be due to the implementation of relatively new biomarkers specific for these diseases. Napsin A has demonstrated higher specificity than TTF-1 for adenocarcinoma.5 Desmoglein-3 is a member of the cadherin adhesion molecule superfamily found in epithelial cells, and is a sensitive and specific marker for SqCC, particularly for the lung. Proteins used in the Pulmotype test were determined by gene expression profiling, and their ability to differentiate between adeno- and squamous cell carcinoma, and were validated in a very large clinical trial.11
EMERGING TECHNOLOGIES MAY INCREASE CDX EFFECTIVENESS
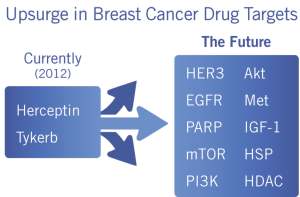
Table 3: New CDx tests may be needed to optimize therapy for next-generation therapies.
This is an exciting time to be at forward-thinking diagnostic laboratories as well as diagnostic manufacturers, as biomarkers, advanced clinical trials based on genomic and proteomic discoveries, and new technology begin to populate the diagnostic landscape. Technologies being introduced into the diagnostics laboratory include mass spectrometry. MALDI TOF-TOF (Matrix-Assisted Laser Desorption Ionization – Time-of-Flight) mass spectrometry is being used to perform imaging of tissues, identifying proteins and peptides within a morphological context with 1-micron precision. Several companies are developing technologies that can track enzyme activity not just of all of the enzymes in a single pathway, but for many pathways simultaneously. What is needed is an inexpensive and easy-to-use method to do this.
Gyrasol Technologies, Lawrence, Kan, a small startup company, is designing a multiplex technology that directly measures enzyme activity for cell-signaling pathways (eg, all of the proteins in Figure 1), and is currently being used in pharmaceutical research. Measures of enzyme activity take into account factors that are missed by genetic tests, such as post-translational modifications, nutritional status, and enzyme cofactor levels, all of which may modulate protein transcription or activity. This takes you one step closer to understanding what is happening biologically in a genetically unstable tumor as it responds to treatment or changes over time. Direct enzyme activity measurement has the potential to measure many different cancer-related signaling pathways simultaneously and rapidly, giving a comprehensive readout of protein activity, assessing responsiveness to many drugs simultaneously. However, one challenge to be overcome if this approach is to become useful in the clinic is that fresh or frozen, rather than fixed, tissue is required for measurement of active enzymes.
A NEW ERA FOR PATHOLOGY AND LAB MEDICINE
Commenting on the pathologist’s response to the emergence of companion diagnostics, Samuel K. Caughron, MD, FCAP, says that for surgical pathologists, companion diagnostics is a component of a much larger trend toward personalized medicine. “Personalized medicine includes advances in genomics and proteomics, as well as sophisticated informatics and digital patient records,” Caughron says. “These will enable access to the vast amounts of patient and test information that can be used to provide care based on that patient’s individual genetic makeup.” Caughron serves as vice chairman for the College of American Pathologists (CAP) Personalized Healthcare Committee, is a member of the Association for Molecular Pathology (AMP) Economic Affairs Committee, and is founding director of the MAWD Molecular Genetic Pathology laboratory, Kansas City, Mo.
Starting with the Human Genome Project—which completed in 2003 and stimulated a resurgence in proteomics research—and combined with a whole series of new technologies for measuring genetic and proteomic markers, a new era of diagnostics is beginning to emerge. With 500 drugs that make use of companion diagnostics currently in the regulatory pipeline, the pathology and laboratory community is being presented with the opportunity to take a stronger leadership role in informing and consulting on patient treatment decisions. Development of novel biomarkers as well as enabling technologies to aid with diagnosis, prognosis, and predictive tests is happening at major universities, reference labs developing and offering novel tests, small startup companies funded by venture capital firms, and large diagnostic companies. As the drug development and diagnostic communities start to come together, as demonstrated by several recent mergers, each side will bring its own distinctive approach. While pharma provides the sheer R&D innovation and resource for development of novel drug targets, these molecules may in turn become biomarkers for disease diagnosis as well as CDx tests as they are validated within the diagnostic laboratory.
Opportunities will abound to make an impact as more companion diagnostics, together with novel, well-validated biomarkers, enter the laboratory. In this climate of change, how does the laboratory and pathology community maintain its relevance? Caughron says that as new tests are developed, the laboratory community can take leadership so that best practices for quality control of personalized medicine and companion diagnostics tests are adopted. “Unlike drug therapies, which stay the same, diagnostic tests can evolve and get better,” he says. “As with HER2 testing, it will be incumbent on us not only to create guidelines for standardizing companion diagnostic tests, but also to work with regulators to establish ways to move diagnostic technology forward to improve technologies when needed.” Pathologists have to somehow avoid becoming just data providers, and instead become integrators of genetic information, working more closely with clinicians, he adds.Greater integration between disciplines will be more and more necessary to make use of an array of treatment advancements that are in development. For example, in breast cancer, there are now only a couple of targeted medications. For HER2 measurement alone, it has taken many years of research and debate to develop the standardized guidelines that are in place today.
Based on medications in advanced clinical trials for breast cancer alone (Table 3), the number of targeted medications may explode in the next few years, giving clinicians additional treatment options and the laboratory many powerful companion diagnostic tests with which to impact patient care.
About half of oncology medications approved recently have a companion diagnostic test associated with them. If this trend continues, pathologists and laboratory directors will have an opportunity to use their expertise in laboratory medicine and standardization to educate and strengthen their partnership with the oncology community. These skills, in combination with the oncologist’s experience in the clinic, will provide much-needed expertise and leadership to integrate many testing algorithms into systematic methods to determine optimal drug therapies and benefit patients.
Mark Cross is a contributing writer for CLP. For more information, contact Editor Judy O’Rourke, .
References
- March 6, 2012, Blog post by Kent Bottles, accessed April 16, 2012. www.hospitalimpact.org/index.php/2012/03/06/4_healthcare_trends_hospital_execs_can_t
- Ries LAG, Young JL, Keel GE, Eisner MP, Lin YD, Horner M-J, eds. SEER Survival Monograph: Cancer Survival Among Adults: US SEER Program, 1988-2001, Patient and Tumor Characteristics. Bethesda, MD: National Cancer Institute, SEER Program; 2007. NIH 07-6215. 9:75.
- Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18-43.
- Johnson DH, Fehrenbacher L, Novotny WF, et al. Randomized phase II trial comparing bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol. 2004;22(11):2184-2191.
- Whithaus K, Fukuoka J, Prihoda TJ, Jagirdar J. Evaluation of napsin A, cytokeratin 5/6, p63, and thyroid transcription factor 1 in adenocarcinoma versus squamous cell carcinoma of the lung. Arch Pathol Lab Med. 2012;136(2):155-162.
- Neal JW. Histology matters: Individualizing Treatment in Non-Small Cell Lung Cancer. Oncologist. 2010;15(1):3-5.
- Edwards SL, Roberts C, McKean ME, et al. Preoperative histological classification of primary lung cancer: accuracy of diagnosis and use of the non-small cell category. J Clin Pathol. 2000;53(7):537-540.
- Mukhopadhyay S, Katzenstein AL. Subclassification of non-small cell lung carcinomas lacking morphologic differentiation on biopsy specimens: Utility of an immunohistochemical panel containing TTF-1, napsin A, p63, and CK5/6. Am J Surg Pathol. 2011;35(1):15-25.
- Tacha D, Yu C, Haas T. TTF-1, Napsin A, p63, TRIM29, Desmoglein-3 and CK5: An Evaluation of Sensitivity and Specificity and Correlation of Tumor Grade for Lung Squamous Cell Carcinoma vs. Lung Adenocarcinoma. Modern Pathology. 2011;24(Supp 1):425A.
- Ring BZ, Seitz RS, Beck RA, et al. A novel five-antibody for subclassification of lung carcinoma. Mod Pathol. 2009;22(8):1032-1043.
- Tacha D, Yu C, Bremer R, Qi W, Haas T. A 6-antibody panel for the classification of lung adenocarcinoma versus squamous cell carcinoma. Appl Immunohistochem Mol Morphol. 2012;20(3):201-207.