One-sentence summary:
MULGONET, a novel multi-omics framework, improves tumor recurrence prediction by integrating gene ontology-guided architecture and attention-based data fusion for interpretable, trans-cancer risk assessment.
Three brief takeaways:
- Overcomes limitations of traditional models by eliminating manual feature selection and capturing pathway-level interactions.
- Hierarchical structure uses over 11,000 Gene Ontology terms to enable broad applicability across cancer types.
- Efficient processing fuses multi-omics data in under 2 hours, outperforming existing methods in both speed and interpretability.
Predicting tumor recurrence at the molecular level emerges as a critical challenge, especially for patients with complex multi-omics profiles. Traditional prognostic models relying on single biomarkers fail to sufficiently capture the interactions between genomic, epigenetic and transcriptomic drivers.
Multi-omics Face Two Key Obstacles
In particular, multi-omics integration face two key obstacles: the reliance on empirical feature selection limiting cross-cancer applicability and the enormous computational complexity in interpreting biological pathways. To that end, in a study published in Fundamental Research by a team from Central South University and Guangxi University, led by Prof. Jianxin Wang and Dr. Wei Lan, the researchers proposed a novel framework that enables precise and interpretable recurrence risk prediction.
“When analyzing multi-omics data from cancers such as bladder and gastric cancer, traditional machine learning models struggle to capture pathway-level interactions,” explains Lan. “Our new framework, MULGONET, overcomes this limitation through two key advances.”
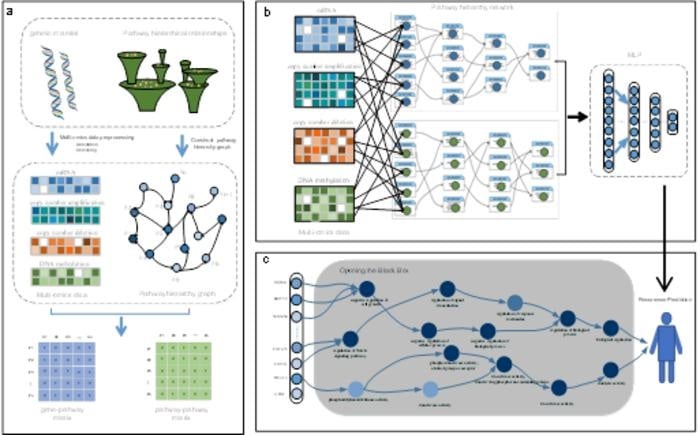
The MULGONET framework
1. Gene ontology guided architecture by building a hierarchical network based on more than 11,000 Gene Ontology (go) terms, MULGONET automatically links genes (such as Cdk6) to biological processes (such as “cell cycle regulation”), eliminating the need for manual feature selection. This enabled the application to trans-cancer, with AUPR scores of 0.774 (bladder cancer), 0.873 (pancreatic cancer) and 0.702 (gastric cancer).
2. Effective multi-omics fusion The model’s attention-based fusion mechanism processes 3D omics data (gene × omics layer × GO term) in less than 2 hours on standard hardware, while existing tools require more than 8 hours.
“Our framework can not only predict recurrence, but also identify driver pathways,” says Lan. “For example, in pancreatic cancer, a high attributable score of Wnt5a in the G protein-coupled receptor signaling pathway is associated with early recurrence.”
The team has opened its framework to accelerate community-driven applications.
“We hope that MULGONET can stimulate new research on the interpretability of multi-omics,” adds Wang.
The study, published in KeAi’s Fundamental Research, directly supports precision oncology by revealing viable targets of Rock1 in metastasis.
Featured Image: Overview of the MULGONET framework. Image: Wei Lan, Zhentao Tang, Haibo Liao et al.