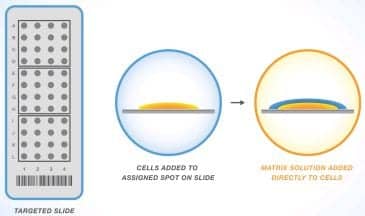
Figure 1. The sample and matrix solution are combined on the target slide.
The microbiology field continues to evolve as organisms become more resistant and difficult to identify. This has led the exploration into new technologies and platforms with the potential to deliver rapid identification—but that also hold other capabilities that allow labs to do more with less equipment and staff. This is critically important for labs that find themselves with strict financial limitations and increasing volumes of work. In the past 5 years, researchers have looked to molecular diagnostics and proteomic technologies for solutions, resulting in labs utilizing technologies new to microbiology. These include polymerase chain reaction (PCR) technology, DNA chips, sequencing technology, mass spectrometry, immunoassays testing, and molecular hybridization probes. These molecular technologies hold the key to keeping up with the increasing number of superbugs that threaten public health.

Nedal Safwat, PhD
is director of product marketing for bioMérieux Inc, Durham, NC.
Accurate molecular diagnostic tools are vital to achieving the kind of personalized medicine that has been promised since the human genome was first mapped in the early days of this century. One platform that holds tremendous potential toward realizing that promise is based on technology first conceived in the mid-1980s with the introduction of the first commercial MALDI-TOF by Shimadzu Corp, Kyoto, Japan. Today, researchers are actively using the technology, known as Matrix-Assisted Laser Desorption Ionization – Time-of-Flight Mass Spectrometry (MALDI-TOF MS) to rapidly identify proteins and large molecules, aiding in genotyping and protein biomarker discovery. For instance, MALDI-TOF MS has been extremely successful in helping to identify various cancer biomarkers. A novel approach took it even one step further in defining microbial organism-specific proteins that have shown a lot of promise for rapid species identification. This ability to deliver fast, accurate results has made MALDI-TOF MS an incredibly versatile tool with uses in biochemistry.
MALDI-TOF MS is currently a research-use-only technology that has been explored in different labs assessing the impact of this technology for routine clinical use in the near future. The speed of the technology is remarkable; it is able to return pathogen identification results in minutes, compared to standard biochemical technique that can take from 6 to 48 hours. This regained time has the potential to enable labs to quickly identify the organism to a species level, leading to starting targeted therapy or change in therapy as much as 24 hours earlier. The future impact will be more effective treatment, faster recovery, and lower cost of treatment due to a targeted therapy. This is especially critical for organisms that are known to be resistant to certain antimicrobials, where rapid identification of the pathogen can lead to potential modification of treatment and taking a more targeted therapy.
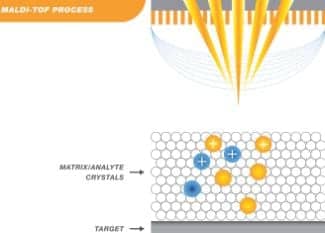
Figure 2. The laser causes the matrix crystals to absorb energy, resulting in the vaporization of proteins, which then accelerate through the Time-of- Flight tube.
LOOKING FROM THE CLINICAL MICROBIOLOGY LAB PERSPECTIVE
Determining whether or not a patient has one of these infections utilizing conventional testing is an extremely time-consuming process. Specimens are streaked, or inoculated, onto media in culture plates and then set aside to culture for anywhere between 18 to 48 hours. Of course, waiting for enough growth on the culture plate is a major bottleneck in the workflow of the lab. If something goes wrong, such as an insufficiently inoculated plate or mixed culture, the process must be repeated. Unfortunately, this is more commonly the rule and not the exception, which means it can often take up to a week to receive accurate test results. The nature of current tests hinder the lab from providing further data, which is needed to provide any further targeted therapy.
Culture-based technology is still considered state-of-the-art in the typical US microbiology lab. This technology is both slow and onerous. Lab scientists have longed for faster technology, which bypasses many of these labor-intensive chores and helps to consolidate testing for multiple pathogen types.
MALDI-TOF MS represents an opportunity to address these most critical areas for improvement. By identifying organisms more quickly, labs will be able to provide hospital clinicians the data they need to more effectively guide therapies. This promises to result in more lives saved and fewer health care dollars going toward longer hospital stays or the treatment of infection-related complications.
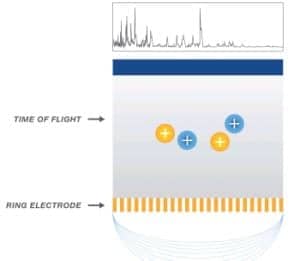
Figure 3. Proteins reach the detector in order of size, smallest to largest, creating the spectra.
MALDI-TOF MS: HOW IT WORKS
MALDI-TOF MS is able to provide a rapid identification of an organism to a species level within minutes utilizing the proteomic fingerprint of the organism tested. MALDI-TOF MS technology allows the separation of proteins based on size and charge, resulting in a specific fingerprint known as spectra, representing the proteins detected and their associated charge to mass ratio. The separation based on charge and size of proteins allows separation of proteins similar in size.
The MALDI-TOF systems have evolved considerably since the first commercial version was introduced by Shimadzu in 1988. The strength of MALDI-TOF MS lies in the ability to generate a protein profile representing a large number of proteins expressed by the specific organism tested. This introduces a high level of protein multiplex testing with results in minutes. The protein profile can be used for microbial identification, detecting specific strains, and detection of antibiotic-resistant markers.
The MALDI process starts by adding the sample to a metal target slide. A matrix solution is then applied to the sample, resulting in formation of crystals (Figure 1).
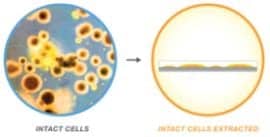
Figure 4. Whole cell analysis is made possible due to the ability to more easily select and analyze intact cells.
The sample is ionized with a laser pulse where the absorption of high energy by the matrix crystals results in vaporization of the proteins that are accelerated by a voltage gradient through the Time-of-Flight tube (Figure 2).
The velocity of the proteins through the Time-of-Flight tube is inversely proportional to the size and charge of the protein (Figure 3). As the proteins separate based on size and charge, smaller proteins reach the detector first, followed by larger proteins, resulting in the creation of a series of peaks named spectra.
The use of mass spectrometry for bacterial identification holds a great deal of promise in terms of allowing hospital labs to process large volumes of samples economically. The speed of results capable with MALDI-TOF MS will work to elevate the importance of the hospital lab, as clinicians will receive pathogen identification very quickly. Faster pathogen identification will be crucial to implementing patient-specific therapy and improving patient outcomes in the future.
FINGERPRINTS OF WHOLE CELLS
Traditional testing of microbes on MALDI-TOF required preparation of the organism through multiple steps to prepare a lysate to be applied to the target slide that was followed by matrix addition. The extraction process is time-consuming and can lead to inconsistencies. The possibility of introducing whole cell analysis was demonstrated through a number of studies1 showing that mass spectra of whole cells of bacteria were identical to mass spectra of extracted samples. The studies revolutionized the approach to testing because now users can analyze intact cells without having to go through multiple extraction steps dealing with traceability and potential contamination (Figures 4 and 5).
DATABASE DEVELOPMENT
The results from the MALDI-TOF MS are quite extensive, providing a series of peaks or mass spectra representing all of the proteins present in the mass range tested. Interpreting the mass spectra has been the focus of scientists for the past decade, with many efforts to build databases composed of known spectra patterns for species of organisms. Interpretation of the mass spectra against a database starts by using a mathematical algorithm to account for significant peaks or proteins. The following step is matching the generated spectra against the database to find a matching pattern. Once a matching pattern is found, then the results are generated with the organism identification.
A new generation of databases has evolved over the past couple of years, taking a new approach that accounts for strain variations, different growth conditions, multiple stages of growth, and other variables that can result in different protein expression. One of the databases that used this approach is the SARAMIS database that was developed using the SuperSpectra approach, which accounts for known variables and builds the specific fingerprint for each species of organisms. Therefore, the software is able to analyze the generated mass spectra and determine the presence of proteins specific to the organism on a genus, family, species, and even strain level. Optimization of existing databases is an ongoing process with a focus on organism groups that require more strain testing, including anaerobes, filamentous fungi, mycobacterium, nocardia, and others.
IMPACT ON THE LAB AND PATIENTS
Once the technology has been approved for clinical use, a lab that adopts the MALDI-TOF MS platform for microbial identification can expect to see several key benefits. In addition to the aforementioned speed of the technology, MALDI-TOF MS provides improved accuracy versus conventional methods via its ability to test more specimens at once in minutes rather than days.
MALDI-TOF will also introduce a single platform that allows the lab to consolidate all of its testing for microbial identification. With its capabilities to identify Gram-negative bacteria, Gram-positive bacteria, fungi, yeast, mycobacterium, and other pathogens, MALDI-TOF represents a single technology that would help eliminate time-consuming methods and improve the way antimicrobial therapy is used.
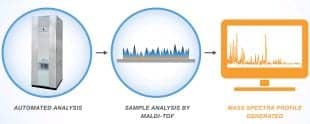
Figure 5. MALDI-TOF allows for rapid ID, as it can take only a few minutes from automated analysis to the generation of the mass spectra.
MALDI-TOF MS shows considerable promise in improving therapy, especially for conditions where the identification of the organism can lead to clear change in antimicrobial therapy. With MALDI-TOF MS, the therapy for specific organisms can me optimized 24 to 48 hours earlier than traditional methods currently used today.
bioMérieux is currently partnering with Shimadzu, a pioneer in the development of the MALDI-TOF MS platform, to integrate the testing capabilities of MALDI-TOF MS and its knowledge base of microbial identification algorithms and databases over the past 12 years. The goal is to apply mass spectrometry for microbial identification once clinical usage is approved, creating the antibiotic susceptibility test on the bioMérieux VITEK® 2 platform. The more labs that are able to provide results to clinicians with the speed and accuracy made possible through MALDI-TOF MS, the fewer the cases of antimicrobial misuse.
Nedal Safwat, PhD, is director of product marketing for bioMérieux Inc, Durham, NC.
Reference
- Shah HN and Gharbia SE, eds. Mass Spectrometry for Microbial Proteomics. John Wiley and Sons; 2010.