Summary:
Researchers at the University of Arizona have developed a low-cost, AI-driven method using label-free optical microscopy to identify cancer phenotypes, potentially making precision medicine more accessible.
Takeaways:
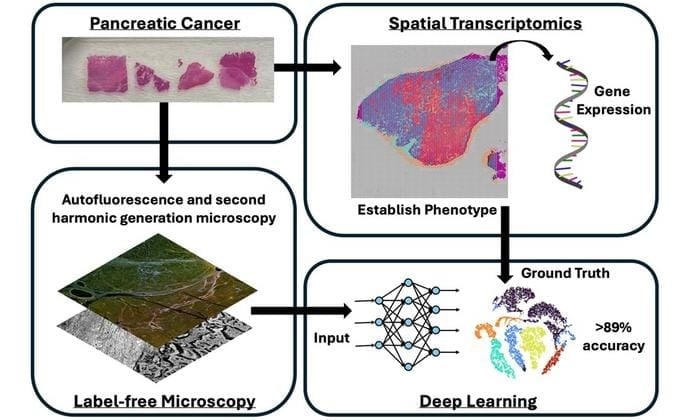
- The method combines spatial transcriptomics and label-free imaging to map gene expression and tissue structure without dyes or genetic tests.
- A deep neural network achieved nearly 90% accuracy in predicting phenotypes from optical images alone, outperforming traditional analysis.
- This innovation could reduce reliance on costly molecular tests, expanding access to precision cancer therapies.
“Precision medicine” has become increasingly popular in the last decade as an avenue for cancer therapy, where treatment strategies are tailored to a specific patient based on the unique characteristics of their disease and their personal background. These unique disease characteristics (called phenotypes) help guide physicians in choosing the most effective treatments.
Using precision medicine can lead to better survival rates and improve the quality of life for patients with cancer. However, while there have been major breakthroughs in using this approach to treat cancer and test new drugs, the tools used to identify disease phenotypes have lagged. Currently, identifying these phenotypes often requires expensive tests, such as those that examine molecular markers, use special stains on tissue samples, or sequence a person’s genetic material. Because of this barrier, many of the potential benefits of precision medicine remain out of reach for many patients.
Recently, a research team based at the University of Arizona developed a faster and more affordable way to identify disease phenotypes in pancreatic cancer. Their work, published in Biophotonics Discovery, describes a new method for disease phenotyping using label-free optical microscopy and artificial intelligence (AI).
Spatial Transciptomics Used to Generate Spatial Maps of Gene Expression
Using a recently developed technology known as spatial transcriptomics, the researchers were able to generate spatial maps of the tissue’s gene expression, allowing them to understand how the disease may behave and to establish phenotypes. The team then performed label-free optical microscopy on the same specimens to form images by measuring natural fluorescence from different biomarkers, as well as measure a response known as second harmonic generation, which is commonly produced by the structural protein collagen. These label-free microscopy images were then co-aligned with the spatial transcriptomic information.
The team then developed an AI algorithm, a deep neural network, which was trained to predict the tissue’s phenotype based only on the label-free optical microscopy images. The approach was able to successfully predict tissue phenotypes to nearly 90 percent accuracy, an exciting finding that demonstrates the promise of label-free microscopy and artificial intelligence for precision medicine applications.
AI Necessary to Aid Identifying Phenotypes
The researchers also showed that classical image analysis methods were not able to extract sufficient information to predict phenotypes, indicating that AI-based methods are necessary to link label-free optical images with characteristics related to underlying disease mechanisms that lead to different phenotypes. This study is also among the first in a new frontier to explore the interface between genetic sequencing and label-free optical imaging methods.
This new method shows that it may be possible to identify disease phenotypes using only light-based imaging and artificial intelligence, without the need for expensive or complex tests. This study marks a significant step forward in applications of optical imaging within precision medicine, which ultimately could make precision medicine more accessible and effective in the future.
For details, see the original Gold Open Access article by S. Guan et al., “Optical phenotyping using label-free microscopy and deep learning,” Biophoton. Discovery 2(3), 035001 (2025) doi: 10.1117/1.BIOS.2.3.035001.
Featured Image: Researchers have developed a new method for pancreatic cancer phenotyping using label-free optical microscopy coupled with a deep learning artificial intelligence algorithm. Using this approach, it was possible to identify tissue phenotypes defined by spatial transcriptomics to over 89% accuracy using label-free microscopy images alone. Image: T. Sawyer (University of Arizona), S. Guan et al., doi 10.1117/1.BIOS.2.3.035001