Monitoring cancer therapy in real time enables physicians to make smarter treatment decisions
By George Karlin-Neumann, PhD
In recent years, the number of treatment options for patients with cancer has increased significantly, and precision medicines are being designed for ever-more-specific patient populations. However, different patients can respond to the same treatment with varying degrees of success, so it is becoming increasingly imperative that physicians have the capacity to accurately predict whether a patient will benefit from a particular therapy.
Nevertheless, our understanding of human biology—especially that of the immune system, genome, and of tumors themselves—is not yet advanced enough to determine with certainty whether therapies, targeted or not, will be effective. So, it’s also critical to monitor patient response.
Surgery is often a first-line treatment option, but it can be risky, and it’s challenging to determine whether surgery will help the patient. By one estimate, one in five patients who underwent elective cancer surgery experienced complications.1 Especially in advanced cancers and cancers that preferentially affect the elderly, surgery can lead to complications such as bleeding, infection, blood clots, and early death—worse outcomes than if the patient had avoided surgery.2 It is therefore critical that physicians assess a patient’s cancer as accurately as possible to determine whether surgery is worth the risk.
For patients who follow surgery with chemotherapy or other adjuvant therapies, the treatment may eventually lose its effectiveness. For example, 90% of chemotherapy failures are due to the emergence of mutations that make the tumor drug resistant.3 This phenomenon can be attributed to several complex factors, such as tumor heterogeneity, the tumor’s ability to evolve, and even the tumor microenvironment, making resistance hard to predict and avoid. Similar factors underlie resistance to other systemic therapies.
Methods for Monitoring Treatment Response
When patients present to a physician with cancer symptoms, it’s often recommended that they undergo some or all of the following diagnostic procedures.
- Computed tomography (CT) or magnetic resonance imaging (MRI) to measure the size and location of tumors.
- Blood tests to assess the presence and level of protein biomarkers.
- Tissue biopsy to assess the tumor’s cellular, molecular, and genetic identity by histology and next-generation sequencing (NGS).
These techniques provide invaluable information about a patient’s initial diagnosis, indicating what type of treatment might be needed based on the size, location, identity, and spread of the cancer. However, once treatment has been initiated, each of these techniques is disadvantageous for helping the physician monitor treatment response and guide subsequent treatment decisions.
Imaging provides only phenotypic information, and the results it yields can be misleading. In patients undergoing immunotherapy, for example, the treatment itself can cause a tumor to grow due to beneficial inflammation, which can cause a physician to falsely believe that a patient’s cancer is worsening. Because of this ‘pseudoprogression,’ which occurs in roughly 9% to 30% of patients, depending on cancer type, use of imaging alone can take months to determine whether an immunotherapy is truly effective.4
Blood tests for protein biomarkers can also help physicians to determine the presence and stage of cancer, but like imaging, they have limited specificity and often lead to inaccurate conclusions. For example, several protein biomarkers commonly used to assess cancer progression, such as prostate-specific antigen for prostate cancer and serum lactate dehydrogenase for melanoma, are unreliable.5,6
Tissue biopsies can provide both phenotypic and genetic information, and in these regards this technique is a more reliable and comprehensive method for profiling a patient’s tumor over time. But tissue biopsies are highly invasive, typically involving a large needle, an endoscope, or surgery. And they can often be risky and painful, and may require anesthesia. So serial monitoring is usually infeasible.7,8 Tissue biopsies also take weeks to return results, taking away valuable time for making an adjustment to a patient’s therapy, and they may not represent the heterogeneity present in a patient’s total tumor burden.
Liquid biopsies provide an alternative means of real-time patient monitoring that is rapidly gaining adoption. The technique offers a less-invasive and potentially more systemic account of tumor variation than is provided by tissue biopsies. Unlike a tissue biopsy, a liquid biopsy only requires a simple blood draw, and can therefore be performed on a serial basis to track a patient’s cancer burden (see the companion article, “Droplet Digital PCR Technology, Step by Step”).
The results of liquid biopsies can be more accurate than those of either imaging or tissue biopsies because of their systemic nature. In a single test, liquid biopsies enable the detection and monitoring of both a phenotypic marker of tumor turnover and highly specific genetic biomarkers for the tumor itself: circulating tumor DNA (ctDNA). Such DNA fragments enter the blood when tumor cells die, and they carry tumor-specific genetic mutations that can be tracked. Levels of ctDNA often correlate with tumor load, and can provide a quantitative measure of tumor progression.9 Finally, ctDNA levels change in response to a therapy’s success or failure, usually within days or weeks, giving oncologists opportunities to rapidly adjust a patient’s treatment regimen.10
Droplet Digital PCR as a Liquid Biopsy Tool
Droplet digital PCR technology has emerged as one of the preferred tools for analyzing ctDNA because of its sensitivity, precision, speed, and reliability (Figure 1). Liquid biopsy can be paired with next-generation sequencing (NGS), which provides a sensitive and comprehensive genetic assessment of a patient’s cancer, and may be best suited for identifying diagnostic biomarkers to guide initial treatment. However, the high cost and long turnaround times associated with NGS make that technique less practical for use in serial monitoring, where only a more restricted set of mutations needs to be followed.
Liquid biopsies based on ddPCR technology are already used in the clinic. They are used in diagnostics for chronic myeloid leukemia, non-small cell lung cancer (NSCLC), and other cancers, demonstrating the reliability and suitability of the technique for use with patient specimens.11 For NSCLC, one commercially available ddPCR liquid biopsy test can detect the major actionable cancer mutations—including single-nucleotide variants, indels, and rearrangements—and deliver results to an oncologist within 72 hours, enabling advance planning of a patient’s treatment to be presented at the first consultation (Figure 2).12
Additionally, numerous clinical studies on bladder cancer, breast cancer, colorectal cancer, melanoma, and other cancers, have shown that a ddPCR-based liquid biopsy can detect and monitor cancer mutations, thereby helping physicians to plan the most favorable treatments for their patients (Figure 3).
A small proof-of-principle study conducted at the University of Leicester showed that a liquid biopsy test based on ddPCR could potentially be used to determine whether patients with malignant pleural mesothelioma might benefit from surgery.13 In the study, researchers identified cancer-associated mutations using whole-exome sequencing and designed a ddPCR liquid biopsy that used as little as 10 ng of cell-free DNA (cfDNA) to assess patients’ chances of surgical success. They found that patients with detectable ctDNA mutations in the blood survived for a significantly shorter amount of time than those without such mutations. The researchers’ finding suggests that this liquid biopsy test could help to stratify patients according to the likelihood that surgery would have a net benefit on their health.
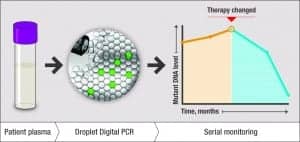
Figure 3. An illustration of ddPCR technology’s ability to track tumor burden by quantifying circulating tumor DNA (ctDNA). A ddPCR liquid biopsy can be administered on a serial basis to monitor ctDNA levels over time, both prior to and in response to therapy.
In addition to predicting the benefit of cancer surgery, ddPCR can also monitor the benefit of remaining on an initial hormone therapy based on the detection of arising resistance mutations. In estrogen-receptor-positive (ER+) breast cancer, the preferred first-line therapy is aromatase-inhibitor hormone therapy. But this type of therapy is not 100% effective, and 25% to 30% of patients experience treatment resistance as ESR1 mutations emerge.14
In an ongoing, large-scale Phase 3 clinical trial at the Institut Curie, researchers are finding that liquid biopsy based on ddPCR technology can detect the acquisition of treatment resistance in ER+ breast cancer patients in response to endocrine therapy.15 Using liquid biopsy tests to serially monitor patients, the researchers were able to witness the emergence of ESR1 mutations linked to endocrine therapy resistance in real time, enabling the team to assign patients to a new treatment group that might offer greater benefit (Figure 4).
When applied for the purpose of tracking treatment resistance, liquid biopsies offer the benefits of speed, reliability, and relative noninvasiveness compared to tissue biopsies. In addition, researchers have found that the technique can, in fact, be more accurate than tissue biopsies for predicting treatment resistance.16 In a Phase 3 clinical trial at the Dana Farber Cancer Institute, researchers tracked mutations in two genes implicated in endocrine therapy resistance in hormone receptor-positive (HR+) metastatic breast cancer, PIK3CA and ESR1. They examined 334 baseline plasma samples and 343 baseline formalin-fixed, paraffin-embedded (FFPE) samples from a total of 558 patients treated with either fulvestrant alone or fulvestrant plus abemaciclib.
The researchers found a correlation between the plasma concentration of mutant PIK3CA and ESR1 and resistance to abemaciclib in plasma ctDNA samples—but not FFPE samples—indicating that ctDNA may be a more reliable predictor of treatment resistance for patients with HR+ metastatic breast cancer.
The Growing Role of ddPCR in the Clinic
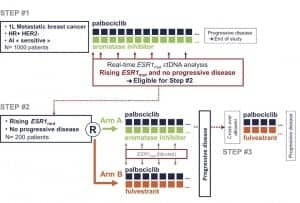
Figure 4. The design of the PADA-1 trial led by Francois-Clement Bidard, MD, PhD, professor of medicine at the University of Paris-Saclay and a research physician in medical oncology at the Institut Curie. Patients were first placed on a palbociclib aromatase inhibitor regimen and were monitored for the onset of ESR1 resistance mutations using a ddPCR liquid biopsy. Half of the nonprogressing patients who exhibited a rise in ESR1 mutations were randomized to an alternative palbociclib-fulvestrant regimen.
As investigators continue to demonstrate the potential of ddPCR-based liquid biopsies for predicting and monitoring patient response to cancer treatments, including surgery and endocrine therapy, such biopsies could very well become a vital component of clinical diagnostic workflows. Liquid biopsies are especially useful after cancer has been diagnosed using imaging and tissue-based characterization. But even in the absence of tissue genotyping, liquid biopsies based on ddPCR technology can be used to noninvasively detect ctDNA as a highly specific marker of tumor subtype and response to treatment.
By measuring a patient’s ctDNA levels, a physician can simultaneously profile a tumor and monitor its severity and progression. From such data, a physician can choose the optimal treatment regimen from the start and, if the patient begins showing signs of progression or treatment resistance, the physician can then adjust the treatment to maximize the potential for a positive outcome. Overall, routine monitoring could improve cancer care, increasing the success rate of treatment, and reducing cancer’s mortality rate.
George Karlin-Neumann, PhD, is director of scientific affairs in the digital biology group at Bio-Rad Laboratories. For further information, contact CLP chief editor Steve Halasey via [email protected].
REFERENCES
- Wong SL, Revels SL, Yin H, et al. Variation in hospital mortality rates with inpatient cancer surgery. Ann Surg. 2015;261(4):632–636; doi: 10.1097/sla.0000000000000690.
- Risks of cancer surgery [online]. Atlanta: American Cancer Society, 2018. Available at: www.cancer.org/treatment/treatments-and-side-effects/treatment-types/surgery/risks-of-cancer-surgery.html. Accessed May 6, 2019.
- Masoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7(3):339–348; doi: 10.15171/apb.2017.041.
- Thrust SC, van den Bent MJ, Smits M. Pseudoprogression of brain tumors. J Magn Reson Imaging. 2018;48(3):571–589; doi: 10.1002/jmri.26171.
- Adhyam M, Gupta AK. A review on the clinical utility of PSA in cancer prostate. Indian J Surg Oncol. 2012;3(2):120–129; doi: 10.1007/s1393-012-0142-6.
- Tsao SCH, Weiss J, Hudson C, et al. Monitoring response to therapy in melanoma by quantifying circulating tumor DNA with droplet digital PCR for BRAF and NRAS mutations. Sci Rep. 2015;5:11198; doi: 10.1038/srep11198.
- Liquid biopsy: using DNA in blood to detect, track, and treat cancer. Bethesda, Md: National Cancer Institute, 2017. Available at: www.cancer.gov/news-events/cancer-currents-blog/2017/liquid-biopsy-detects-treats-cancer. Accessed May 6, 2019.
- Types of biopsies used to look for cancer [online]. Atlanta: American Cancer Society, 2015. Available at: www.cancer.org/treatment/understanding-your-diagnosis/tests/testing-biopsy-and-cytology-specimens-for-cancer/biopsy-types.html. Accessed May 6, 2019.
- Lee Y, Park S, Kim WS, et al. Correlation between progression-free survival, tumor burden, and circulating tumor DNA in the initial diagnosis of advanced-stage EGFR-mutated non-small cell lung cancer. Thorac Cancer. 2018;9(9):1104–1110; doi: 10.1111/1759-7714.12793.
- Hiltermann TJN, Miedema A, Hijmerjing-Kappelle L, et al. Very early molecular marker of tumor response to PD-1 inhibition in plasma of patients with advanced non-small cell lung cancer (NSCLC). J Clin Oncol. 2017;35(15 suppl):3030–3030; doi: 10.1200/jco.2017.35.15_suppl.3030.
- Mellert H, Alexander K, Jackson L, et al. Early feasibility and development of multiplexed, single-reaction assay for ALK, ROS1, and RET novel ddPCR RNA fusions [poster, online]. Poster presented at the annual meeting of the American Association for Cancer Research, Washington, DC, April 1–5, 2017. Available at: www.biodesix.com/early-feasibility-development-multiplexed-single-reaction-assay-alk-ros1-ret-novel-ddpcr-rna-fusions. Accessed May 27, 2019.
- Evans R. Blood test is ready for clinical decisionmaking: ddPCR detection of EGFR and KRAS mutations in ctDNA and EML4-ALK in circulating RNA show high sensitivity, specificity, and concordance [online]. Boulder, Colo: Biodesix, 2016. Available at: www.biodesix.com/ddpcr-blood-test-for-egfp-and-kras-mutations. Accessed May 27, 2019.
- Martinson LJ, Sharkey AJ, Dawson AG, et al. Personalized circulating tumor DNA profiling in malignant pleural mesothelioma [abstract 1349/18, online]. Presentation at the annual meeting of the American Association for Cancer Research, Atlanta, March 29–April 3, 2019. Available at: www.abstractsonline.com/pp8/#!/6812/presentation/5024. Accessed May 27, 2019.
- Clatot F, Augusto L, Di Fiore F. ESR1 mutations in breast cancer. Aging (Albany NY). 2017;9(1):3–4; doi: 10.18632/aging.101165.
- Bidard FC. Clinical utility trials for CTC and ctDNA in ER+ advanced breast cancer [abstract SY31-02, online]. Presentation at the annual meeting of the American Association for Cancer Research, Atlanta, March 29–April 3, 2019. Available at: www.abstractsonline.com/pp8/#!/6812/presentation/7958. Accessed May 27, 2019.
- Tolaney SM, Toi M, Neven P, et al. Clinical significance of PIK3CA and ESR1 mutations in ctDNA and FFPE samples from the Monarch 2 study of abemaciclib plus fulvestrant [abstract 4458, online]. Presentation at the annual meeting of the American Association for Cancer Research, Atlanta, March 29–April 3, 2019. Available at: www.abstractsonline.com/pp8/#!/6812/presentation/4767. Accessed May 27, 2019.





Great Blog. Thanks for sharing the information about Liquid Biopsies Guide Improved Cancer Treatment. This blog is highly informative and helpful for all internet users. Keep sharing this type of info.
My mother was diagnosed with cancer last month, which is why we’re currently looking for treatments. Well, you made a pretty good point that blood tests and the level of protein biomarkers will be needed. Thank you for also informing us about the importance of conducting a ddPCR.